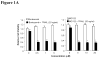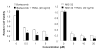Proteasome inhibitors-mediated TRAIL resensitization and Bik accumulation - PubMed (original) (raw)
Proteasome inhibitors-mediated TRAIL resensitization and Bik accumulation
Hongbo Zhu et al. Cancer Biol Ther. 2005 Jul.
Abstract
Proteasome inhibitors can resensitize cells that are resistant to tumor necrosis factor-related apoptotic-inducing ligand (TRAIL)-mediated apoptosis. However, the underlying mechanisms of this effect are unclear. To characterize the mechanisms of interaction between proteasome inhibitors and TRAIL protein, we evaluated the effects of combined treatment with the proteasome inhibitors bortezomib and MG132 and TRAIL protein on two TRAIL-resistant human colon cancer cell lines, DLD1-TRAIL/R and LOVO-TRAIL/R. Both bortezomib and MG132 in combination with TRAIL enhanced apoptotosis induction in these cells, as evidenced by enhanced cleavage of caspases 8, 9, and 3, Bid, poly(ADP-ribose) polymerase and by the release of cytochrome C and Smac. Subsequent studies showed that combined treatment with bortezomib or MG132 resulted in an increase of death receptor (DR) 5 and Bik at protein levels but had no effects on protein levels of DR4, Bax, Bak, Bcl-2, Bcl-XL or Flice-inhibitory protein (FLIP). Moreover, c-Jun N-terminal kinase (JNK) is activated by these proteasome inhibitors. Blocking JNK activation with the JNK inhibitor SP600125 attenuated DR5 increase, but enhancement of apoptosis induction and increase of Bik protein were not affected. However, bortezomib-mediated TRAIL sensitization was partially blocked by using siRNA to knockdown Bik. Thus, our data suggests that accumulation of Bik may be critical for proteasome inhibitor-mediated resensitization of TRAIL.
Figures
FIG. 1
Combined effects of proteasome inhibitors and TRAIL protein in TRAIL-resistant DLD1-TRAIL/R cells. DLD1-TRAIL/R cells were treated with bortezomib or MG132 for 2 h, followed by 20 ng/ml of TRAIL protein for 4 h. (A) Cell viability was determined by XTT assay and (B) proportion of apoptotic cells determined by FACS analysis. (C). Cell viability 24 h after addition of TRAIL protein. Each assay was performed in quadruplicate. The data presented are means + SD. * P < 0.05 compared with the proteasome inhibitor alone.
FIG. 1
Combined effects of proteasome inhibitors and TRAIL protein in TRAIL-resistant DLD1-TRAIL/R cells. DLD1-TRAIL/R cells were treated with bortezomib or MG132 for 2 h, followed by 20 ng/ml of TRAIL protein for 4 h. (A) Cell viability was determined by XTT assay and (B) proportion of apoptotic cells determined by FACS analysis. (C). Cell viability 24 h after addition of TRAIL protein. Each assay was performed in quadruplicate. The data presented are means + SD. * P < 0.05 compared with the proteasome inhibitor alone.
FIG. 1
Combined effects of proteasome inhibitors and TRAIL protein in TRAIL-resistant DLD1-TRAIL/R cells. DLD1-TRAIL/R cells were treated with bortezomib or MG132 for 2 h, followed by 20 ng/ml of TRAIL protein for 4 h. (A) Cell viability was determined by XTT assay and (B) proportion of apoptotic cells determined by FACS analysis. (C). Cell viability 24 h after addition of TRAIL protein. Each assay was performed in quadruplicate. The data presented are means + SD. * P < 0.05 compared with the proteasome inhibitor alone.
FIG. 2
Combined effect of proteasome inhibitors and TRAIL protein in TRAIL-resistant LOVO-TRAIL/R cells. LOVO-TRAIL/R cells were treated with bortezomib or MG132 for 2 h, followed by 20 ng/ml of TRAIL protein for 24 h. Cell viability was then determined by XTT assay. The data presented are mean + SD of triplicate assays. * P < 0.05 compared with the proteasome inhibitor alone.
FIG. 3
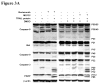
Apoptosis profiles of DLD1-TRAIL/R cells treated with bortezomib (1 μM) or MG132 (5 μM) for 2 h, followed by 20 ng/ml of TRAIL protein for 4 h. Cell lysates were then subjected to Western blot analysis. (A) cleavage of caspases. (B) Release of cytochrome C and Smac. The mitochondrial fraction (Mito) was used as a positive control. (C) Caspase activation 24 h after addition of TRAIL protein. The data presented were from one of two independent experiments with similar results.
FIG. 3
Apoptosis profiles of DLD1-TRAIL/R cells treated with bortezomib (1 μM) or MG132 (5 μM) for 2 h, followed by 20 ng/ml of TRAIL protein for 4 h. Cell lysates were then subjected to Western blot analysis. (A) cleavage of caspases. (B) Release of cytochrome C and Smac. The mitochondrial fraction (Mito) was used as a positive control. (C) Caspase activation 24 h after addition of TRAIL protein. The data presented were from one of two independent experiments with similar results.
FIG. 3
Apoptosis profiles of DLD1-TRAIL/R cells treated with bortezomib (1 μM) or MG132 (5 μM) for 2 h, followed by 20 ng/ml of TRAIL protein for 4 h. Cell lysates were then subjected to Western blot analysis. (A) cleavage of caspases. (B) Release of cytochrome C and Smac. The mitochondrial fraction (Mito) was used as a positive control. (C) Caspase activation 24 h after addition of TRAIL protein. The data presented were from one of two independent experiments with similar results.
FIG. 4
DR5 and Bik upregulation by proteasome inhibitors. DLD1-TRAIL/R TRAIL-resistant cells treated with bortezomib (1 μM) or MG132 (5 μM) for 2 h followed by 20 ng/ml of TRAIL protein for 4 h. The cell lysates were then subjected to Western blot analysis. The data presented are from one of two independent experiments with similar results.
FIG. 5
JNK pathway activation by proteasome inhibitors. (A). JNK activation detected in TRAIL-resistant DLD1-TRAIL/R cells treated as described in the Fig. 4. The data presented are from one of two independent experiments with similar results. (B) Effect of JNK inhibitor. DLD1-TRAIL/R cells were treated with 50-μM in SP600125 for 2 h, followed by the combination treatment mentioned in Fig. 4. The cell lysates were then subjected to Western blot analysis. The data presented one of two independent experiments with similar results. (C) Apoptosis in cells treated as in B and determined by FACS analysis. The data represented means + SD of a triplicate assays. (D) Effect of Bik siRNA on proteasome inhibitor mediated TRAIL re-sensitization. DLD1-TRAIL/R cells were treated with 100 nM of Bik siRNA or Luciferase siRNA as indicated. 24 hours later, 1 μM of bortezomib was added for 2 hours, followed by 20ng/ml of TRAIL protein for another 4 hours. Apoptosis was then determined by FACS. The value represents mean + SD of a triplicate assay. * represents P<0.05.
FIG. 5
JNK pathway activation by proteasome inhibitors. (A). JNK activation detected in TRAIL-resistant DLD1-TRAIL/R cells treated as described in the Fig. 4. The data presented are from one of two independent experiments with similar results. (B) Effect of JNK inhibitor. DLD1-TRAIL/R cells were treated with 50-μM in SP600125 for 2 h, followed by the combination treatment mentioned in Fig. 4. The cell lysates were then subjected to Western blot analysis. The data presented one of two independent experiments with similar results. (C) Apoptosis in cells treated as in B and determined by FACS analysis. The data represented means + SD of a triplicate assays. (D) Effect of Bik siRNA on proteasome inhibitor mediated TRAIL re-sensitization. DLD1-TRAIL/R cells were treated with 100 nM of Bik siRNA or Luciferase siRNA as indicated. 24 hours later, 1 μM of bortezomib was added for 2 hours, followed by 20ng/ml of TRAIL protein for another 4 hours. Apoptosis was then determined by FACS. The value represents mean + SD of a triplicate assay. * represents P<0.05.
FIG. 5
JNK pathway activation by proteasome inhibitors. (A). JNK activation detected in TRAIL-resistant DLD1-TRAIL/R cells treated as described in the Fig. 4. The data presented are from one of two independent experiments with similar results. (B) Effect of JNK inhibitor. DLD1-TRAIL/R cells were treated with 50-μM in SP600125 for 2 h, followed by the combination treatment mentioned in Fig. 4. The cell lysates were then subjected to Western blot analysis. The data presented one of two independent experiments with similar results. (C) Apoptosis in cells treated as in B and determined by FACS analysis. The data represented means + SD of a triplicate assays. (D) Effect of Bik siRNA on proteasome inhibitor mediated TRAIL re-sensitization. DLD1-TRAIL/R cells were treated with 100 nM of Bik siRNA or Luciferase siRNA as indicated. 24 hours later, 1 μM of bortezomib was added for 2 hours, followed by 20ng/ml of TRAIL protein for another 4 hours. Apoptosis was then determined by FACS. The value represents mean + SD of a triplicate assay. * represents P<0.05.
FIG. 5
JNK pathway activation by proteasome inhibitors. (A). JNK activation detected in TRAIL-resistant DLD1-TRAIL/R cells treated as described in the Fig. 4. The data presented are from one of two independent experiments with similar results. (B) Effect of JNK inhibitor. DLD1-TRAIL/R cells were treated with 50-μM in SP600125 for 2 h, followed by the combination treatment mentioned in Fig. 4. The cell lysates were then subjected to Western blot analysis. The data presented one of two independent experiments with similar results. (C) Apoptosis in cells treated as in B and determined by FACS analysis. The data represented means + SD of a triplicate assays. (D) Effect of Bik siRNA on proteasome inhibitor mediated TRAIL re-sensitization. DLD1-TRAIL/R cells were treated with 100 nM of Bik siRNA or Luciferase siRNA as indicated. 24 hours later, 1 μM of bortezomib was added for 2 hours, followed by 20ng/ml of TRAIL protein for another 4 hours. Apoptosis was then determined by FACS. The value represents mean + SD of a triplicate assay. * represents P<0.05.
Similar articles
- The proteasome inhibitor MG132 potentiates TRAIL receptor agonist-induced apoptosis by stabilizing tBid and Bik in human head and neck squamous cell carcinoma cells.
Sung ES, Park KJ, Choi HJ, Kim CH, Kim YS. Sung ES, et al. Exp Cell Res. 2012 Aug 1;318(13):1564-76. doi: 10.1016/j.yexcr.2012.04.003. Epub 2012 Apr 10. Exp Cell Res. 2012. PMID: 22513214 - The proteasome inhibitor bortezomib sensitizes cells to killing by death receptor ligand TRAIL via BH3-only proteins Bik and Bim.
Nikrad M, Johnson T, Puthalalath H, Coultas L, Adams J, Kraft AS. Nikrad M, et al. Mol Cancer Ther. 2005 Mar;4(3):443-9. doi: 10.1158/1535-7163.MCT-04-0260. Mol Cancer Ther. 2005. PMID: 15767553 - The TRAIL apoptotic pathway mediates proteasome inhibitor induced apoptosis in primary chronic lymphocytic leukemia cells.
Kabore AF, Sun J, Hu X, McCrea K, Johnston JB, Gibson SB. Kabore AF, et al. Apoptosis. 2006 Jul;11(7):1175-93. doi: 10.1007/s10495-006-8048-9. Apoptosis. 2006. PMID: 16699949 - Mechanisms of resistance to TRAIL-induced apoptosis in cancer.
Zhang L, Fang B. Zhang L, et al. Cancer Gene Ther. 2005 Mar;12(3):228-37. doi: 10.1038/sj.cgt.7700792. Cancer Gene Ther. 2005. PMID: 15550937 Review. - Combining proteasome inhibition with TNF-related apoptosis-inducing ligand (Apo2L/TRAIL) for cancer therapy.
Sayers TJ, Murphy WJ. Sayers TJ, et al. Cancer Immunol Immunother. 2006 Jan;55(1):76-84. doi: 10.1007/s00262-005-0676-3. Epub 2005 Oct 27. Cancer Immunol Immunother. 2006. PMID: 15864587 Free PMC article. Review.
Cited by
- Bortezomib synergizes TRAIL-induced apoptosis in gastric cancer cells.
Liu J, Qu XJ, Xu L, Zang Y, Qu JL, Hou KZ, Liu YP. Liu J, et al. Dig Dis Sci. 2010 Dec;55(12):3361-8. doi: 10.1007/s10620-010-1191-8. Dig Dis Sci. 2010. PMID: 20393880 - The ubiquitin-proteasome system: opportunities for therapeutic intervention in solid tumors.
Johnson DE. Johnson DE. Endocr Relat Cancer. 2015 Feb;22(1):T1-17. doi: 10.1530/ERC-14-0005. Epub 2014 Mar 21. Endocr Relat Cancer. 2015. PMID: 24659480 Free PMC article. Review. - A cell-based high-throughput screen to identify synergistic TRAIL sensitizers.
Booth NL, Sayers TJ, Brooks AD, Thomas CL, Jacobsen K, Goncharova EI, McMahon JB, Henrich CJ. Booth NL, et al. Cancer Immunol Immunother. 2009 Aug;58(8):1229-44. doi: 10.1007/s00262-008-0637-8. Epub 2008 Dec 17. Cancer Immunol Immunother. 2009. PMID: 19089423 Free PMC article. - BIK/NBK gene as potential marker of prognostic and therapeutic target in breast cancer patients.
López-Muñoz E, Hernández-Zarco A, García-Hernández N, Alvarado-Cabrero I, Zarco-Espinosa G, Salamanca-Gómez F, Arenas-Aranda D. López-Muñoz E, et al. Clin Transl Oncol. 2012 Aug;14(8):586-91. doi: 10.1007/s12094-012-0845-8. Epub 2012 Jul 11. Clin Transl Oncol. 2012. PMID: 22855140 - Manipulating the apoptotic pathway: potential therapeutics for cancer patients.
Bates DJ, Lewis LD. Bates DJ, et al. Br J Clin Pharmacol. 2013 Sep;76(3):381-95. doi: 10.1111/bcp.12193. Br J Clin Pharmacol. 2013. PMID: 23782006 Free PMC article. Review.
References
- Eggert A, Grotzer MA, Zuzak TJ, Wiewrodt BR, Ho R, et al. Resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Cancer Res. 2001;61:1314–1319. - PubMed
- Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000;60:4315–4319. - PubMed
- Zhang L, Gu J, Lin T, Huang X, Roth JA, Fang B. Mechanisms involved in development of resistance to adenovirus-mediated proapoptotic gene therapy in DLD1 human colon cancer cell line. Gene Ther. 2002;9:1262–1270. - PubMed
- Kim K, Takimoto R, Dicker DT, Chen Y, Gazitt Y, El-Deiry WS. Enhanced TRAIL sensitivity by p53 overexpression in human cancer but not normal cell lines. Inter J Oncol. 2001;18:241–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA092487/CA/NCI NIH HHS/United States
- CA16672/CA/NCI NIH HHS/United States
- CA 092487-01A1/CA/NCI NIH HHS/United States
- R01 CA098582/CA/NCI NIH HHS/United States
- CA 098582-01A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous