Clioquinol down-regulates mutant huntingtin expression in vitro and mitigates pathology in a Huntington's disease mouse model - PubMed (original) (raw)
Clioquinol down-regulates mutant huntingtin expression in vitro and mitigates pathology in a Huntington's disease mouse model
Trent Nguyen et al. Proc Natl Acad Sci U S A. 2005.
Abstract
In investigating the role of metal ions in the pathogenesis of Huntington's disease, we examined the effects of clioquinol, a metal-binding compound currently in clinical trials for Alzheimer's disease treatment, on mutant huntingtin-expressing cells. We found that PC12 cells expressing polyglutamine-expanded huntingtin exon 1 accumulated less mutant protein and showed decreased cell death when treated with clioquinol. This effect was polyglutamine-length-specific and did not alter mRNA levels or protein degradation rates. Clioquinol treatment of transgenic Huntington's mice (R6/2) improved behavioral and pathologic phenotypes, including decreased huntingtin aggregate accumulation, decreased striatal atrophy, improved rotarod performance, reduction of weight loss, normalization of blood glucose and insulin levels, and extension of lifespan. Our results suggest that clioquinol is a candidate therapy for Huntington's disease and other polyglutamine-expansion diseases.
Figures
Fig. 1.
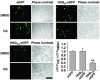
CQ down-regulates expanded polyQ-egfp fluorescence in vitro. Bars and error bars represent mean + SEM. n ≥ 3 for each data set. Fluorescence microscopy and quantitation of EGFP-, Httexon1-Q25-egfp-, and Httexon1-Q103-_egfp_-expressing cells. (Bar, 100 μm.) **, P < 0.03.
Fig. 2.
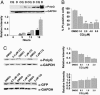
CQ down-regulates polyQ protein expression and decreases cell death in vitro. Bars and error bars represent mean + SEM. n ≥ 3 for each data set. Western data are normalized to the GAPDH signal. (A) Western blotting and quantitation of Httexon1-Q103-egfp (polyQ Ab) in HEK293 cell lysates; times posttransfection as indicated. (B) Viability (propidium iodide staining) of Httexon1-Q103-egfp cells treated with CQ. *, P ≤ 0.03; **, P < 0.002 vs. vehicle control. (C) Western blotting and quantitation of Httexon1-Q103-egfp (polyQ Ab) and EGFP (GFP Ab) in cell lysates. ***, P = 0.02.
Fig. 3.
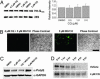
CQ does not affect polyQ mRNA levels or protein degradation. Bars and error bars represent mean + SEM. n ≥ 3 for each data set. (A) Northern blotting and quantitation of Httexon1-Q103-egfp transcripts. (B) Fluorescence microscopy of mutant protein accumulation in Httexon1-Q103-_egfp-_expressing cells in presence of MG132 ± CQ. (Bar, 100 μm.) (C) Western analysis of cells treated with MG132 and CQ. (D) Pulse–chase analysis of Httexon1-Q103-egfp turnover in the presence and absence of CQ.
Fig. 4.
CQ inhibits mutant Htt aggregate accumulation in vivo. Bars and error bars represent mean + SEM. (A) Representative Western blotting and quantitation of high molecular weight mutant Htt in whole-brain homogenates from 11-week-old R6/2 transgenic mice. Wt, wild-type; Tg, vehicle-treated R6/2; CQ, CQ-treated R6/2. n = 3. *, P = 0.01. (B) Inhibition of neuronal intranuclear inclusion formation by treatment with CQ. Representative images of striatum and cortex from 11-week-old mice; wild-type, R6/2-vehicle treated, or R6/2-CQ treated, as indicated. Htt aggregates were visualized by using EM48 Ab. Results were similar for six or more animals in each treatment group. (Bar, 50 μm.)
Fig. 5.
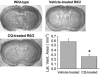
CQ decreases polyQ-mediated pathology in vivo. Bars and error bars represent mean + SEM. Representative images of the cerebrum of 11-week-old mice and quantitation of lateral ventricle areas, showing decreased striatal atrophy in 11-week-old CQ-treated R6/2 mice. n ≥ 6 for each treatment group. *, P < 0.001 vs. vehicle-treated mice.
Fig. 6.
CQ positively effects behavior, weight, and survival in vivo. Bars and error bars represent mean + SEM. (A) Illustration of clasping behavior. (Left) Wild-type mouse. (Right) Ten-week-old R6/2 mouse exhibiting clasping. (B) Clasping score at 10 weeks of age in CQ and vehicle-treated R6/2. n = 8. *, P < 0.0004 vs. vehicle. (C) Rotarod testing of CQ and vehicle-treated R6/2. Bars and error bars represent mean + SEM. Black bars, vehicle-treated; gray bars, CQ-treated. n ≥ 6 for each group. *, P < 0.03; **, P < 0.001 vs. vehicle-treated animals. (D) Body weight over time of wild-type (filled circles), vehicle-treated R6/2 (open circles), and CQ-treated R6/2 (filled triangles); symbols and bars represent mean ± SEM. n ≥ 6 for each group, *, P < 0.01 vs. vehicle-treated animals by Student's t test; also, P < 0.0001 overall, for CQ-treated vs. vehicle-treated animals by ANOVA with post hoc Bonferroni/Dunn testing. (E) Kaplan–Meier analysis of R6/2 lifespan, vehicle treated (closed circles) vs. CQ treated (open circles). n = 5 per group. P = 0.0018 by log-rank Mantel–Cox test.
Similar articles
- Olesoxime suppresses calpain activation and mutant huntingtin fragmentation in the BACHD rat.
Clemens LE, Weber JJ, Wlodkowski TT, Yu-Taeger L, Michaud M, Calaminus C, Eckert SH, Gaca J, Weiss A, Magg JC, Jansson EK, Eckert GP, Pichler BJ, Bordet T, Pruss RM, Riess O, Nguyen HP. Clemens LE, et al. Brain. 2015 Dec;138(Pt 12):3632-53. doi: 10.1093/brain/awv290. Epub 2015 Oct 21. Brain. 2015. PMID: 26490331 - Mutant huntingtin and glycogen synthase kinase 3-beta accumulate in neuronal lipid rafts of a presymptomatic knock-in mouse model of Huntington's disease.
Valencia A, Reeves PB, Sapp E, Li X, Alexander J, Kegel KB, Chase K, Aronin N, DiFiglia M. Valencia A, et al. J Neurosci Res. 2010 Jan;88(1):179-90. doi: 10.1002/jnr.22184. J Neurosci Res. 2010. PMID: 19642201 - Huntington's disease does not appear to increase the risk of diabetes mellitus.
Boesgaard TW, Nielsen TT, Josefsen K, Hansen T, Jørgensen T, Pedersen O, Nørremølle A, Nielsen JE, Hasholt L. Boesgaard TW, et al. J Neuroendocrinol. 2009 Sep;21(9):770-6. doi: 10.1111/j.1365-2826.2009.01898.x. Epub 2009 Jul 7. J Neuroendocrinol. 2009. PMID: 19602103 - Molecular aspects of Huntington's disease.
Walling HW, Baldassare JJ, Westfall TC. Walling HW, et al. J Neurosci Res. 1998 Nov 1;54(3):301-8. doi: 10.1002/(SICI)1097-4547(19981101)54:3<301::AID-JNR1>3.0.CO;2-W. J Neurosci Res. 1998. PMID: 9819135 Review. - Huntingtin-protein interactions and the pathogenesis of Huntington's disease.
Li SH, Li XJ. Li SH, et al. Trends Genet. 2004 Mar;20(3):146-54. doi: 10.1016/j.tig.2004.01.008. Trends Genet. 2004. PMID: 15036808 Review.
Cited by
- Trans-synaptic zinc mobilization improves social interaction in two mouse models of autism through NMDAR activation.
Lee EJ, Lee H, Huang TN, Chung C, Shin W, Kim K, Koh JY, Hsueh YP, Kim E. Lee EJ, et al. Nat Commun. 2015 May 18;6:7168. doi: 10.1038/ncomms8168. Nat Commun. 2015. PMID: 25981743 Free PMC article. - Therapeutic Strategies in Huntington's Disease.
Kanazawa I. Kanazawa I. J Clin Neurol. 2006 Dec;2(4):213-24. doi: 10.3988/jcn.2006.2.4.213. Epub 2006 Dec 20. J Clin Neurol. 2006. PMID: 20396523 Free PMC article. - Ubiquitin-Proteasome System in Neurodegenerative Disorders.
Rao G, Croft B, Teng C, Awasthi V. Rao G, et al. J Drug Metab Toxicol. 2015;6(4):187. doi: 10.4172/2157-7609.1000187. Epub 2015 Aug 13. J Drug Metab Toxicol. 2015. PMID: 30761219 Free PMC article. - Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Ali MG, Wahba HM, Igelmann S, Cyr N, Ferbeyre G, Omichinski JG. Ali MG, et al. Autophagy. 2024 Sep;20(9):1948-1967. doi: 10.1080/15548627.2024.2353443. Epub 2024 May 27. Autophagy. 2024. PMID: 38726830 - Navigating the Chemical Space of Multitarget-Directed Ligands: From Hybrids to Fragments in Alzheimer's Disease.
Prati F, Cavalli A, Bolognesi ML. Prati F, et al. Molecules. 2016 Apr 8;21(4):466. doi: 10.3390/molecules21040466. Molecules. 2016. PMID: 27070562 Free PMC article. Review.
References
- Bates, G. P., Harper, P. S. & Jones, L. (2002) Huntington's Disease (Oxford Univ. Press, Oxford, U.K.).
- Snell, R. G., MacMillan, J. C., Cheadle, J. P., Fenton, I., Lazarou, L. P., Davies, P., MacDonald, M. E., Gusella, J. F., Harper, P. S. & Shaw, D. J. (1993) Nat. Genet. 4, 393-397. - PubMed
- Ambrose, C. M., Duyao, M. P., Barnes, G., Bates, G. P., Lin, C. S., Srinidhi, J., Baxendale, S., Hummerich, H., Lehrach, H., Altherr, M., et al. (1994) Somat. Cell Mol. Genet. 20, 27-38. - PubMed
- Gauthier, L. R., Charrin, B. C., Borrell-Pages, M., Dompierre, J. P., Rangone, H., Cordelieres, F. P., De Mey, J., MacDonald, M. E., Lessmann, V., Humbert, S., et al. (2004) Cell 118, 127-138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical