Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis - PubMed (original) (raw)
Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis
Lesley F Drynan et al. EMBO J. 2005.
Abstract
Chromosomal translocations are primary events in tumorigenesis. Those involving the mixed lineage leukaemia (MLL) gene are found in various guises and it is unclear whether MLL fusions can affect haematopoietic differentiation. We have used a model in which chromosomal translocations are generated in mice de novo by Cre-loxP-mediated recombination (translocator mice) to compare the functionally relevant haematopoietic cell contexts for Mll fusions, namely pluripotent stem cells, semicommitted progenitors or committed cells. Translocations between Mll and Enl or Af9 cause myeloid neoplasias, initiating in pluripotent stem cells or multipotent myeloid progenitors. However, while Mll-Enl translocations can also cause leukaemia from T-cell progenitors, no tumours arose with Mll-Af9 translocations in the T-cell compartment. Furthermore, Mll-Enl translocations in T-cell progenitors can cause lineage reassignment into myeloid tumours. Therefore, a permissive cellular environment is required for oncogenicity of Mll-associated translocations and Mll fusions can influence haematopoietic lineage commitment.
Figures
Figure 1
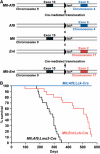
Incidence of leukaemias in Mll translocator mice. (A) Diagram of Mll, Enl and Af9 targeted alleles in translocator mice (Collins et al, 2000; Forster et al, 2003). LoxP sites were introduced, by homologous recombination in ES cells, into introns of mouse Mll, Enl or Af9, corresponding to the human introns where chromosomal translocations are typically found. Specific cells in mice carrying loxP sites in both Mll and Enl or Mll and Af9 genes can undergo chromosomal translocations, mediated by Cre recombinase, to create fusion genes analogous to those of human leukaemias, as indicated. (B) Survival curves for translocator mice. Cohorts of 20 translocator mice expressing Cre under the control of Lck or Lmo2 promoters (McCormack et al, 2003) were analysed over a period of about 1.5 years.
Figure 2
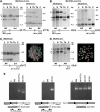
Haematological malignancies in Mll translocators have chromosomal translocations. Mll; Enl; Lck-Cre (A, C) or Mll; Af9; Lmo2-Cre (B, D) tumours were examined for the presence of chromosomal translocations by filter hybridisation and by FISH. For the former, DNA samples are designated as follows: C: control C57BL/6 tail or CCB ES DNA; Ta: tail; Th: thymus; S: spleen; L: liver. For each panel, the spleen, liver and thymus DNAs were prepared from Mll; Enl; Lck-Cre or Mll; Af9; Lmo2-Cre mice (overlined samples). Hybridising bands correspond to targeted (T), germline (G) or chromosomal translocation (CT) alleles. Hybridisation probes from either side of the translocation junction, namely 5′ and 3′ Mll probes and a 3′ Enl probe (A and C, respectively) or 5′ and 3′ Mll probes and a 3′ Af9 probe (B and D, respectively), were used to detect rearrangements. For FISH analysis (C, D), metaphase spreads were made with cells from the spleen and painted with whole chromosome paints for chromosome 9 (green) and chromosome 4 (Af9) or 17 (Enl) (red). Painted chromosomes from an Mll; Enl; Lck-Cre (C) or Mll; Af9; Lmo2-Cre spleen (D) are shown. (E) RT–PCR analysis of _Mll_-fusion mRNA. Cell lines were established from Mll-AF9 knock-in mice (Corral et al, 1996; Dobson et al, 1999), Mll; Enl; Lmo2-Cre translocators (Forster et al, 2003) and Mll; Af9; Lmo2-Cre translocators (this paper). RNA was made and converted to cDNA for RT–PCR amplification with primers spanning junctions of Mll and Enl or Af9 RNA sequences as indicated. Total bone marrow RNA from a wild-type mouse was used as a negative control for cDNA lacking Mll-fusion sequences and RT–PCR with actin primers was used for quality control of cDNA. C: no template (H2O) control; BM: bone marrow; ki: knock-in.
Figure 3
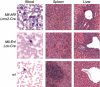
Histopathology of tumours from Mll-Enl and Mll-Af9 translocator mice. Tissues were fixed in 10% formalin and wax embedded prior to generation of histological sections. The sections were stained with H&E and photographed at × 40 magnification. May–Grünwald–Giemsa (MGG)-stained blood films and sections shown are from an Mll; Af9; Lmo2-Cre translocator mouse (top row), from an Mll; Enl; Lck-Cre translocator mouse (middle row) or from a C57BL/6 wild-type mouse (bottom row).
Figure 4
Cell surface profiles of leukaemias vary in different translocator mice. Single-cell suspensions from splenic tumours of Mll; Enl; Lck-Cre or Mll; Af9; Lmo2-Cre translocator mice were compared by flow cytometry with splenic leucocytes from a control (Mll; Enl) mouse lacking the Cre allele. Surface proteins were labelled with fluorescent antibodies detecting CD8 plus CD4 (T-cell markers) or Mac-1 plus Gr1 (myeloid markers).
Figure 5
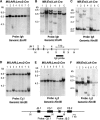
Rearranged antigen receptor genes in tumours from Mll; Enl; Lck-Cre translocator mice. Genomic DNA from splenic tumours of Mll; Af9; Lmo2-Cre (A, D, E) or Mll; Enl; Lck-Cre (B, C, F) translocator mice were analysed by filter hybridisation using an Igh, Tcr C_β_1 or Tcr J_β_2 probe. For hybridisation of the DNAs with the Igh probes, digestions were either with _Eco_RI (A, B), which will detect all joining events to the whole Ig JH region, or with _Hin_dIII (C). Hybridisation of TCR probes was with _Hin_dIII-digested DNA. V-D-J or D-J joins were detected with C_β_1 or J_β_2 probes. Partial restriction maps of the JH or C_β_1–J_β_2 genomic regions are shown, indicating the location of the probe fragments. H: _Hin_dIII; X: _Xba_I; R: _Eco_RI.
Figure 6
Chromosomal translocations are present in Mll; Af9; Lck-Cre translocators by postnatal day 8 and Mll-Af9 expressed at 6 months. (A–C) Bone marrow (BM) and thymus (Thy) genomic DNA was prepared from 8-day-old pups (five littermates for each genotype) from Mll; Af9; Lck-Cre or Mll; Enl; Lck-Cre translocators. PCR was performed for 70 cycles using nested primers to amplify either (A) Mll plus Enl (MG1+EG1, followed by MN+EN; Forster et al, 2003) or (B, C) Mll plus Af9 primers (1a+1b, followed by 2a+2b; Collins et al, 2000). Positive controls were performed on DNA from Mll; Af9 or Mll; Enl translocator splenic tumours (indicated as AML) using 35 cycles of amplification, and negative controls were performed, using 70 cycles of amplification, on DNA prepared from spleens of P8 litter mates of Mll; Af9 or Mll; Enl mice without the Cre gene (indicated as Cre−). Quality control of DNA was determined by PCR amplification of the Lmo2 gene with primers B5 and B3 (McCormack et al, 2003) using 35 cycles of amplification. Primer locations are diagrammatically shown. Bands corresponding to amplified products are indicated as follows: ME: Mll-Enl junction; MA: Mll-Af9 junction; L2: Lmo2 gene product. (D) Litters of Mll; Af9; Lck-Cre mice, including Mll; Af9 controls lacking Cre gene, were established and at 3 or 6 months, RNA was prepared from the spleen, thymus, kidney and bone marrow (latter at 6 months only). RT–PCR was carried out with primers from exon 10 of Mll or exon 9 of Af9 to detect RNA products of the Mll-Af9 translocated fusion allele. The double-headed arrow indicates the gel band corresponding to the product of this translocated allele. Mll-Af9 fusion mRNA transcript was present in the spleen and thymus at 3 months and in addition detected in these tissues and in bone marrow at 6 months of translocator mice. M: φX174 _Hae_III-digested DNA markers; +ve: RT–PCR product from an Mll-Af9 leukaemia produced in Mll; Af9; Lmo2-Cre mice; −ve: RT–PCR in the absence of template; BM: bone marrow.
Figure 7
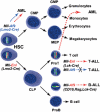
Summary of Mll fusions in haematopoiesis and tumorigenesis. When chromosomal translocations are generated de novo in Mll-Af9 or Mll-Enl translocator mice, haematopoietic malignancies arise if translocations are mediated via Cre expression with either Lmo2-Cre (Mll-Af9 or Mll-Enl) or Lck-Cre (Mll-Enl only) alleles. No tumours arose when Mll-Af9 chromosomal translocations were induced by Lck-Cre expression, even though both chromosomal translocations and Mll-Af9 fusion mRNA could be detected. Thus, when Mll translocations are induced in the pluripotent stem cells or in the multipotent myeloid progenitors using Lmo2-Cre, myeloid malignancies arise (this paper and Forster et al, 2003). A more complex situation was observed when the chromosomal translocations between Mll and Enl genes were induced by _Lck-Cre_-controlled expression of Cre recombinase. In this case, either T-cell malignancies or myeloid leukaemias appeared; however, in both types, the leukaemia progenitors had arisen from cells that had expressed functional Rag V-D-J recombinase. Thus, the myeloid malignancies of the Mll; Enl; Lck-Cre translocator mice appear to arise from cells that have undergone a lineage reassignment from a semicommitted, Rag+ lymphoid cell to a myeloid cell. This effect may occur by reprogramming the transcriptome of cells with the chromosomal translocations to realigning differentiation into myeloid from lymphoid status. HSC: haematopoietic stem cells; CMP: common myeloid progenitors; GMP: granulocyte–monocyte precursors; MEP: megakaryocyte–erythrocyte precursors; CLP: common lymphoid progenitors.
Similar articles
- MLL-ENL-mediated leukemia initiation at the interface of lymphoid commitment.
Ugale A, Säwén P, Dudenhöffer-Pfeifer M, Wahlestedt M, Norddahl GL, Bryder D. Ugale A, et al. Oncogene. 2017 Jun 1;36(22):3207-3212. doi: 10.1038/onc.2016.470. Epub 2017 Jan 9. Oncogene. 2017. PMID: 28068328 - Leukaemia lineage specification caused by cell-specific Mll-Enl translocations.
Cano F, Drynan LF, Pannell R, Rabbitts TH. Cano F, et al. Oncogene. 2008 Mar 20;27(13):1945-50. doi: 10.1038/sj.onc.1210818. Epub 2007 Oct 1. Oncogene. 2008. PMID: 17906700 - MLL-AF9 leukemia stem cells: hardwired or taking cues from the microenvironment?
Muntean AG, Hess JL. Muntean AG, et al. Cancer Cell. 2008 Jun;13(6):465-7. doi: 10.1016/j.ccr.2008.05.012. Cancer Cell. 2008. PMID: 18538728 - Learning from mouse models of MLL fusion gene-driven acute leukemia.
Schwaller J. Schwaller J. Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194550. doi: 10.1016/j.bbagrm.2020.194550. Epub 2020 Apr 19. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32320749 Review. - Transcriptional activation by MLL fusion proteins in leukemogenesis.
Yokoyama A. Yokoyama A. Exp Hematol. 2017 Feb;46:21-30. doi: 10.1016/j.exphem.2016.10.014. Epub 2016 Nov 16. Exp Hematol. 2017. PMID: 27865805 Review.
Cited by
- ENL: structure, function, and roles in hematopoiesis and acute myeloid leukemia.
Zhou J, Ng Y, Chng WJ. Zhou J, et al. Cell Mol Life Sci. 2018 Nov;75(21):3931-3941. doi: 10.1007/s00018-018-2895-8. Epub 2018 Jul 31. Cell Mol Life Sci. 2018. PMID: 30066088 Free PMC article. Review. - Chromosomal Translocation Formation Is Sufficient to Produce Fusion Circular RNAs Specific to Patient Tumor Cells.
Babin L, Piganeau M, Renouf B, Lamribet K, Thirant C, Deriano L, Mercher T, Giovannangeli C, Brunet EC. Babin L, et al. iScience. 2018 Jul 27;5:19-29. doi: 10.1016/j.isci.2018.06.007. Epub 2018 Jun 19. iScience. 2018. PMID: 30240643 Free PMC article. - Human MLL-AF9 Overexpression Induces Aberrant Hematopoietic Expansion in Zebrafish.
Tan J, Zhao L, Wang G, Li T, Li D, Xu Q, Chen X, Shang Z, Wang J, Zhou J. Tan J, et al. Biomed Res Int. 2018 May 30;2018:6705842. doi: 10.1155/2018/6705842. eCollection 2018. Biomed Res Int. 2018. PMID: 30003105 Free PMC article. - The "Never-Ending" Mouse Models for MLL-Rearranged Acute Leukemia Are Still Teaching Us.
Ottersbach K, Sanjuan-Pla A, Torres-Ruíz R, Bueno C, Velasco-Hernández T, Menendez P. Ottersbach K, et al. Hemasphere. 2018 Jun 19;2(4):e57. doi: 10.1097/HS9.0000000000000057. eCollection 2018 Aug. Hemasphere. 2018. PMID: 31723783 Free PMC article. No abstract available. - MLL-ENL-mediated leukemia initiation at the interface of lymphoid commitment.
Ugale A, Säwén P, Dudenhöffer-Pfeifer M, Wahlestedt M, Norddahl GL, Bryder D. Ugale A, et al. Oncogene. 2017 Jun 1;36(22):3207-3212. doi: 10.1038/onc.2016.470. Epub 2017 Jan 9. Oncogene. 2017. PMID: 28068328
References
- Adriaansen HJ, Soeting PW, Wolvers-Tettero IL, van Dongen JJ (1991) Immunoglobulin and T-cell receptor gene rearrangements in acute non-lymphocytic leukemias. Analysis of 54 cases and a review of the literature. Leukemia 5: 744–751 - PubMed
- Ayton PM, Cleary ML (2001) Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene Rev 20: 5695–5707 - PubMed
- Boehm T, Werle A, Drahousky D (1987a) Immunoglobulin heavy chain and T-cell receptor α and β chain gene rearrangements in acute myeloid leukaemias. Mol Biol Med 4: 51–62 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases