Characterization of Dicer-deficient murine embryonic stem cells - PubMed (original) (raw)
Comparative Study
. 2005 Aug 23;102(34):12135-40.
doi: 10.1073/pnas.0505479102. Epub 2005 Aug 12.
Affiliations
- PMID: 16099834
- PMCID: PMC1185572
- DOI: 10.1073/pnas.0505479102
Comparative Study
Characterization of Dicer-deficient murine embryonic stem cells
Elizabeth P Murchison et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Dicer is an RNase III-family nuclease that initiates RNA interference (RNAi) and related phenomena by generation of the small RNAs that determine the specificity of these gene silencing pathways. We have previously shown that Dicer is essential for mammalian development, with Dicer-deficient mice dying at embryonic day 7.5 with a lack of detectable multipotent stem cells. To permit a more detailed investigation of the biological roles of Dicer, we have generated embryonic stem cell lines in which their single Dicer gene can be conditionally inactivated. As expected, Dicer loss compromises maturation of microRNAs and leads to a defect in gene silencing triggered by long dsRNAs. However, the absence of Dicer does not affect the ability of small interfering RNAs to repress gene expression. Of interest, Dicer loss does compromise the proliferation of ES cells, possibly rationalizing the phenotype previously observed in Dicer-null animals. Dicer loss also affects the abundance of transcripts from mammalian centromeres but does so without a pronounced affect on histone modification status at pericentric repeats or methylation of centromeric DNA. These studies provide a conditional model of RNAi deficiency in mammals that will permit the dissection of the biological roles of the RNAi machinery in cultured mammalian cells.
Figures
Fig. 1.
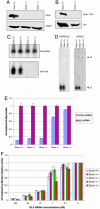
Conditional Dicer ES cells. (A) Dicer domain structure. Dicer floxed allele was created by knocking in a floxed cassette, flanking the RNaseIII domain-encoding exons 22 and 23 with loxP sites. (B) ES cells that are heterozygous for a Dicer-null allele (created by deletion of an RNaseIII domain) have previously been described (1). The introduction of the Dicer floxed allele into these ES cells created Dicer floxed ES cells. Treatment with Cre leads to excision of the majority of both RNaseIII domains, creating a second nonfunctional allele. The four different Dicer alleles (wild-type, null, conditional but not excised, and conditional excised) can be distinguished by Southern blotting. (C) A Southern blot used to confirm the genotypes of the cell lines used in this study is shown.
Fig. 2.
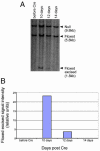
Dicer-null cells show proliferation defects. (A) Dicer floxed ES cells were transiently transfected with a Cre-GFP expression plasmid and sorted for GFP-positive cells, and the population was analyzed at different times after Cre expression by Southern blotting. The Dicerflox(excised)/null population was lost from the mixed population at 10-15 days after excision. (B) The relative representation of the Dicerflox(excised)/null versus the Dicerflox/null cells in the population was quantified by image analysis of the gel shown in A.
Fig. 3.
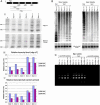
Dicer-null ES cells do not process dsRNA or pre-miRNAs but are able to respond to siRNAs. No full-length Dicer protein can be detected in whole-cell lysates of Dicer-deficient cells. (A) Western blotting using an antibody recognizing the RNaseIII domains of Dicer. (B) Western blotting using an antibody recognizing the PAZ domain of Dicer. Dicer-null ES cells fail to process pre-miRNAs into mature miRNAs. Total RNA extracted from floxed/null and floxed excised/null ES cells was probed with 32P-labeled oligonucleotides complementary to miR-19b (with equal loading indicated by blotting for U6 snRNA) (C) or miR-293 and miR-292-as (D). (E) Dicer-null ES cells fail to initiate gene silencing in response to dsRNA. dsRNAs, 500 nt in length, corresponding to either firefly luciferase or GFP were introduced into ES cells along with firefly and Renilla luciferase reporters. (F) Standard dual luciferase assays were performed on ES cells transfected with siRNAs.
Fig. 4.
Changes in centromeric RNA levels in Dicer-null cells but no change in histone modification status or cytosine methylation in Dicer-null cells. (A) RT-PCR was used to quantify centromere-derived transcripts. (B) Southern blotting was used to determine the methylation status of the major and minor satellite repeats. (C) Antibodies directed against trimethyl histone H3 lysine 9 were used for chromatin immunoprecipitations at the major satellite repeats. Immunoprecipitate (with or without antibody) and input were amplified with primers specific to major satellite repeats with 20 cycles. Along with Dicer wild-type, heterozygous, and null are shown Suv39h1 and h2 double null mouse embryonic fibroblasts together with wild-type mouse embryonic fibroblast controls.
Similar articles
- Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing.
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Kanellopoulou C, et al. Genes Dev. 2005 Feb 15;19(4):489-501. doi: 10.1101/gad.1248505. Genes Dev. 2005. PMID: 15713842 Free PMC article. - Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference.
Nicholson RH, Nicholson AW. Nicholson RH, et al. Mamm Genome. 2002 Feb;13(2):67-73. doi: 10.1007/s00335-001-2119-6. Mamm Genome. 2002. PMID: 11889553 - Critical roles for Dicer in the female germline.
Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Murchison EP, et al. Genes Dev. 2007 Mar 15;21(6):682-93. doi: 10.1101/gad.1521307. Genes Dev. 2007. PMID: 17369401 Free PMC article. - [Advance on Dicer gene and its role in female reproduction].
Li P, Zhu WJ. Li P, et al. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011 Jun;28(3):275-8. doi: 10.3760/cma.j.issn.1003-9406.2011.03.008. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011. PMID: 21644222 Review. Chinese. - [Great potential of small RNAs: RNA interference and microRNA].
Vázquez-Ortiz G, Piña-Sánchez P, Salcedo M. Vázquez-Ortiz G, et al. Rev Invest Clin. 2006 Jul-Aug;58(4):335-49. Rev Invest Clin. 2006. PMID: 17146945 Review. Spanish.
Cited by
- Normal DNA methylation dynamics in DICER1-deficient mouse embryonic stem cells.
Ip J, Canham P, Choo KH, Inaba Y, Jacobs SA, Kalitsis P, Mattiske DM, Ng J, Saffery R, Wong NC, Wong LH, Mann JR. Ip J, et al. PLoS Genet. 2012 Sep;8(9):e1002919. doi: 10.1371/journal.pgen.1002919. Epub 2012 Sep 6. PLoS Genet. 2012. PMID: 22969435 Free PMC article. - siRNA-mediated gene knockdown via electroporation in hydrozoan jellyfish embryos.
Masuda-Ozawa T, Fujita S, Nakamura R, Watanabe H, Kuranaga E, Nakajima YI. Masuda-Ozawa T, et al. Sci Rep. 2022 Sep 30;12(1):16049. doi: 10.1038/s41598-022-20476-1. Sci Rep. 2022. PMID: 36180523 Free PMC article. - miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency.
Cao Y, Guo WT, Tian S, He X, Wang XW, Liu X, Gu KL, Ma X, Huang D, Hu L, Cai Y, Zhang H, Wang Y, Gao P. Cao Y, et al. EMBO J. 2015 Mar 4;34(5):609-23. doi: 10.15252/embj.201490441. Epub 2015 Jan 20. EMBO J. 2015. PMID: 25603933 Free PMC article. - Chromatin measurements reveal contributions of synthesis and decay to steady-state mRNA levels.
Tippmann SC, Ivanek R, Gaidatzis D, Schöler A, Hoerner L, van Nimwegen E, Stadler PF, Stadler MB, Schübeler D. Tippmann SC, et al. Mol Syst Biol. 2012 Jul 17;8:593. doi: 10.1038/msb.2012.23. Mol Syst Biol. 2012. PMID: 22806141 Free PMC article. - Non-canonical microRNAs miR-320 and miR-702 promote proliferation in Dgcr8-deficient embryonic stem cells.
Kim BM, Choi MY. Kim BM, et al. Biochem Biophys Res Commun. 2012 Sep 21;426(2):183-9. doi: 10.1016/j.bbrc.2012.08.058. Epub 2012 Aug 19. Biochem Biophys Res Commun. 2012. PMID: 22925886 Free PMC article.
References
- Bernstein, E., Kim, S. Y., Carmell, M. A., Murchison, E. P., Alcorn, H., Li, M. Z., Mills, A. A., Elledge, S. J., Anderson, K. V. & Hannon, G. J. (2003) Nat. Genet. 35, 215-217. - PubMed
- Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. - PubMed
- Hannon, G. J. (2002) Nature 418, 244-251. - PubMed
- Carmell, M. A. & Hannon, G. J. (2004) Nat. Struct. Mol. Biol. 11, 214-218. - PubMed
- Siolas, D., Lerner, C., Burchard, J., Ge, W., Linsley, P. S., Paddison, P. J., Hannon, G. J. & Cleary, M. A. (2005) Nat. Biotechnol. 23, 227-231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials