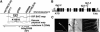The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis - PubMed (original) (raw)
The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis
Piers A Hemsley et al. Plant Cell. 2005 Sep.
Abstract
TIP GROWTH DEFECTIVE1 (TIP1) of Arabidopsis thaliana affects cell growth throughout the plant and has a particularly strong effect on root hair growth. We have identified TIP1 by map-based cloning and complementation of the mutant phenotype. TIP1 encodes an ankyrin repeat protein with a DHHC Cys-rich domain that is expressed in roots, leaves, inflorescence stems, and floral tissue. Two homologues of TIP1 in yeast (Saccharomyces cerevisiae) and human (Homo sapiens) have been shown to have S-acyl transferase (also known as palmitoyl transferase) activity. S-acylation is a reversible hydrophobic protein modification that offers swift, flexible control of protein hydrophobicity and affects protein association with membranes, signal transduction, and vesicle trafficking within cells. We show that TIP1 binds the acyl group palmitate, that it can rescue the morphological, temperature sensitivity, and yeast casein kinase2 localization defects of the yeast S-acyl transferase mutant akr1Delta, and that inhibition of acylation in wild-type Arabidopsis roots reproduces the Tip1- mutant phenotype. Our results demonstrate that S-acylation is essential for normal plant cell growth and identify a plant S-acyl transferase, an essential research tool if we are to understand how this important, reversible lipid modification operates in plant cells.
Figures
Figure 1.
Map-Based Cloning of TIP1. (A) Map location of tip1. The tip1-2 mutation mapped between g4111 and R89998 on chromosome 5. Two artificial chromosome clones, F5P1 and K19B10, both complemented the tip1-2 mutant phenotype (data not shown). IGF, Institut für Genbiologische Forschung Berlin. (B) Structure of TIP1. Introns are shown as lines and exons as black boxes. Two white boxes indicate extra exon sequence present in TIP1 cDNA but incorrectly annotated in public databases. tip1-1 carries a G/A mutation and disrupts the splice site between exon 10 and intron 10. _tip1_-2 carries a Tyr/stop mutation at amino acid 41. tip1-3 harbors a SALK T-DNA insertion in the 3′ end of exon 11. UTR, untranslated region. (C) Biolistic complementation of tip1-2 with At5g20350. Left, fluorescence image of a tip1-2 root cobombarded with Pro35S:_TIP1_cDNA and Pro35S:GFP showing a single transformed root hair expressing GFP. Center, bright-field image of the same root showing mutant hairs (black arrowhead) and a transformed hair with wild-type morphology (white arrowhead). Right, stably transformed tip1-2 plant expressing Pro35S:TIP1 showing long, unbranched root hairs. Bar = 500 μm.
Figure 2.
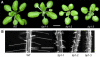
tip1-3 Has a Strong Tip1− Phenotype. (A) tip1-3 has compact, dwarf vegetative growth similar to that of tip1-2. Plants were grown in long days at 18°C and are shown at the eight-leaf stage. Bar = 10 mm. (B) tip1-3 has a strong root hair phenotype similar to the short branched hairs observed on tip1-2 mutant roots. Roots are shown 5 d after germination. Bar = 400 μm.
Figure 3.
TIP1 Shows Homology with Other Proteins from Yeast and Mammals. The greatest homology is observed around the region of the DHHC motif (indicated by asterisks). TIP1 (Arabidopsis), HIP14 (human), AKR1 (yeast), ERF2 (yeast), GODZ (mouse). TIP1 shows greater homology with HIP14, ERF2, and GODZ within the DHHC-CRD than to AKR1p. Residues that are identical in three or more of the five sequences are shaded in black. Conservative substitutions at an amino acid position that occur in three or more of the five sequences are shaded in gray.
Figure 4.
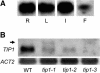
RNA Gel Blot Analysis of TIP1 Expression in Wild-Type and Mutant Plants. (A) TIP1 is expressed throughout the plant. R, root; L, leaf; I, inflorescence; F, flower. (B) The Tip1− mutants all show reduced levels of transcript compared with the wild type. The arrowhead indicates a second larger transcript in tip1-1. No transcript is detectable in tip1-3.
Figure 5.
TIP1 Complements _akr1_Δ Mutant Yeast. _akr1_Δ yeast cells show decreased growth at 30°C compared with 25°C on solid medium, show aberrant morphology in liquid medium, and fail to target YCK2p to the plasma membrane. The temperature-sensitive phenotypes of yeast deficient in AKR1 can be complemented by the addition of TIP1. (A) TIP1 restores wild-type growth to yeast grown on solid medium at 30°C. Single colonies of equal size were streaked onto complete selective medium and incubated at 25 or 30°C. Genotypes are as illustrated in the scheme at bottom. (B) TIP1 but not TIP1 C401A restores the morphology of _akr1_Δ yeast cells grown at 30°C in liquid culture. Liquid cultures were grown to mid-log phase in selective medium at 25 or 30°C before imaging. Bar = 10 μm. (C) TIP1 is capable of targeting YCK2:GFP to the plasma membrane. YCK2p requires AKR1p for correct localization to the plasma membrane in wild-type yeast, and the _akr1_Δ mutant shows a diffuse cytoplasmic localization of YCK2:GFP. TIP1 is capable of substituting for AKR1p in targeting YCK2:GFP to the plasma membrane and requires Cys-401 to do so. Bar = 5 μm.
Figure 6.
Inhibition of Acylation with 2-Bromopalmitate Phenocopies the Tip1− Phenotype. (A) Growing roots were treated with 2-bromopalmitate solution or mock-treated with solvent. The white arrows denote the region of root growth and root hair expansion in the presence of 2-bromopalmitate over the subsequent 6 h before imaging. Bar = 500 μm. (B) Magnified images of tip1-2, mock-treated wild-type, and treated wild-type roots show that 10 μM 2-bromopalmitate treatment produces a root hair phenotype comparable to that of tip1-2. Bar = 400 μm.
Figure 7.
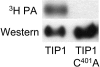
TIP1 Is Labeled with [3H]Palmitic Acid in Yeast. _S_-acyl transferases can be labeled with [3H]palmitic acid (3H PA) in vivo. TIP1 carrying a C401A mutation within the DHHC active site motif fails to label with [3H]palmitic acid. TIP1 and TIP1 C401A were expressed as C-terminal FLAG epitope tag fusions under the GAL1 promoter.
Similar articles
- A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis.
Qi B, Doughty J, Hooley R. Qi B, et al. New Phytol. 2013 Oct;200(2):444-456. doi: 10.1111/nph.12385. Epub 2013 Jun 25. New Phytol. 2013. PMID: 23795888 Free PMC article. - PLURIPETALA mediates ROP2 localization and stability in parallel to SCN1 but synergistically with TIP1 in root hairs.
Chai S, Ge FR, Feng QN, Li S, Zhang Y. Chai S, et al. Plant J. 2016 Jun;86(5):413-25. doi: 10.1111/tpj.13179. Plant J. 2016. PMID: 27037800 - Protein palmitoylation is critical for the polar growth of root hairs in Arabidopsis.
Zhang YL, Li E, Feng QN, Zhao XY, Ge FR, Zhang Y, Li S. Zhang YL, et al. BMC Plant Biol. 2015 Feb 13;15:50. doi: 10.1186/s12870-015-0441-5. BMC Plant Biol. 2015. PMID: 25849075 Free PMC article. - Systematic screening for palmitoyl transferase activity of the DHHC protein family in mammalian cells.
Fukata Y, Iwanaga T, Fukata M. Fukata Y, et al. Methods. 2006 Oct;40(2):177-82. doi: 10.1016/j.ymeth.2006.05.015. Methods. 2006. PMID: 17012030 Review. - Protein palmitoylation by a family of DHHC protein S-acyltransferases.
Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Mitchell DA, et al. J Lipid Res. 2006 Jun;47(6):1118-27. doi: 10.1194/jlr.R600007-JLR200. Epub 2006 Apr 1. J Lipid Res. 2006. PMID: 16582420 Review.
Cited by
- Interaction of OsRopGEF3 Protein With OsRac3 to Regulate Root Hair Elongation and Reactive Oxygen Species Formation in Rice (Oryza sativa).
Kim EJ, Hong WJ, Tun W, An G, Kim ST, Kim YJ, Jung KH. Kim EJ, et al. Front Plant Sci. 2021 May 25;12:661352. doi: 10.3389/fpls.2021.661352. eCollection 2021. Front Plant Sci. 2021. PMID: 34113363 Free PMC article. - Tip growth defective1 interacts with cellulose synthase A3 to regulate cellulose biosynthesis in Arabidopsis.
Zhang L, Thapa Magar MS, Wang Y, Cheng Y. Zhang L, et al. Plant Mol Biol. 2022 Sep;110(1-2):1-12. doi: 10.1007/s11103-022-01283-8. Epub 2022 May 29. Plant Mol Biol. 2022. PMID: 35644016 - Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95.
Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, Tani H, Iwanari H, Mochizuki Y, Kodama T, Matsuura Y, Bredt DS, Hamakubo T, Fukata M. Noritake J, et al. J Cell Biol. 2009 Jul 13;186(1):147-60. doi: 10.1083/jcb.200903101. J Cell Biol. 2009. PMID: 19596852 Free PMC article. - Progress toward Understanding Protein S-acylation: Prospective in Plants.
Li Y, Qi B. Li Y, et al. Front Plant Sci. 2017 Mar 24;8:346. doi: 10.3389/fpls.2017.00346. eCollection 2017. Front Plant Sci. 2017. PMID: 28392791 Free PMC article. Review. - Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family.
Batistic O. Batistic O. Plant Physiol. 2012 Nov;160(3):1597-612. doi: 10.1104/pp.112.203968. Epub 2012 Sep 11. Plant Physiol. 2012. PMID: 22968831 Free PMC article.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
- Babu, P., Deschenes, R.J., and Robinson, L.C. (2004). Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J. Biol. Chem. 279, 27138–27147. - PubMed
- Bagnat, M., and Simons, K. (2002). Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 383, 1475–1480. - PubMed
- Baker, K.E., and Parker, R. (2004). Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16, 293–299. - PubMed
- Böhme, K., Li, Y., Charlot, F., Grierson, C., Marrocco, K., Okada, K., Laloue, M., and Nogue, F. (2004). The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 40, 686–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases