Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors - PubMed (original) (raw)
Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors
Frédéric Clotman et al. Genes Dev. 2005.
Abstract
During liver development, hepatocytes and biliary cells differentiate from common progenitors called hepatoblasts. The factors that control hepatoblast fate decision are unknown. Here we report that a gradient of activin/TGFbeta signaling controls hepatoblast differentiation. High activin/TGFbeta signaling is required near the portal vein for differentiation of biliary cells. The Onecut transcription factors HNF-6 and OC-2 inhibit activin/TGFbeta signaling in the parenchyma, and this allows normal hepatocyte differentiation. In the absence of Onecut factors, the shape of the activin/TGFbeta gradient is perturbed and the hepatoblasts differentiate into hybrid cells that display characteristics of both hepatocytes and biliary cells. Thus, a gradient of activin/TGFbeta signaling modulated by Onecut factors is required to segregate the hepatocytic and the biliary lineages.
Figures
Figure 1.
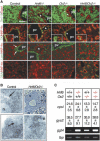
HNF-6 and OC-2 are required to segregate the hepatocytic and biliary lineages. (A) In the absence of HNF-6 and OC-2, the hepatoblasts give rise to hybrid cells coexpressing hepatoblast/hepatocyte and biliary markers. Markers of epithelial cells (Ecad, E-cadherin), erythroid cells (Ter119), hepatoblasts and hepatocytes (HNF-4α), or biliary cells ([CK] cytokeratin; [Lam] laminin; [DBA] Dolichos biflorus agglutinin) were detected on E15.5 liver sections. (Insets) Higher magnifications of CK/HNF-4α labelings. (pv) Portal vein; (arrows) biliary basal lamina; (asterisks) biliary cysts in _Hnf6_–/– livers; (arrowheads) dilations within the ductal plate in _Oc2_–/– livers. Bar, 100 μm. (B) The hybrid cells in _Hnf6/Oc2_–/– livers have morphological characteristics of hepatocytes. For Hnf6/Oc2_–/– liver at E15.5, a semithin section (upper right) shows hybrid cells surrounded by hematopoietic cells; in hybrid cells, a well-developed rough endoplasmic reticulum (lower right), abundant mitochondria, and a bile canaliculus (lower left, arrow) were observed by electron microscopy. (n) Nucleus; (arrows) bile canaliculi. (C) The hepatocyte markers carbamoyl phosphate synthase I (cpsI) and glycogen synthase 2 (gys2) are expressed in Onecut knockout livers, as shown by real-time RT–PCR quantification, [(mRNA copy number)/(β-actin mRNA copy number)] × 103 (mean ± SEM). The expression of the biliary marker γ_-glutamyl transpeptidase 4 (ggt4) is increased in Onecut knockout livers, as shown by semiquantitative RT–PCR with TATA-binding protein gene (tbp) as control.
Figure 2.
HNF-6 and OC-2 inhibit activin/TGFβ signaling. (A) HNF-6 and OC-2 control the expression of components of the activin/TGFβ signaling pathway. Real-time RT–PCR for TGF_β_-receptor type II (tbrII), α_2-macroglobulin_ (a2m) or follistatin at E12.5, expressed as [(mRNA copy number)/(β-actin mRNA copy number)] × 103 (mean ± SEM). (B,C) Activin/TGFβ signaling occurs as a gradient in E12.5 liver and is increased in the absence of Onecut factors. (B) Hnf6+/– Oc2+/– mice were crossbred with transgenic mice harboring an activin/TGFβ reporter consisting of 12 Smad-binding sites upstream of EGFP (CAGA12/GFP). The dotted lines delineate the branches of the portal vein. The inset shows background fluorescence of a section from a nontransgenic liver. (pv) Portal vein. Bar, 100 μm. (C) GFP fluorescence profile from the portal vein to the parenchyma in control CAGA12/GFP liver (blue tracing) and in _Hnf6/Oc2_–/– CAGA12/GFP liver (red tracing). In the parenchyma, basal fluorescence levels correspond to hematopoietic cells, and higher levels to epithelial cells. The dotted lines show the shape of GFP fluorescence gradients in the epithelial cells. “Ductal plate” refers to the area where the ductal plate will form. (pv) Portal vein.
Figure 3.
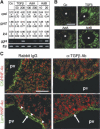
Activin/TGFβ signaling induces biliary differentiation. (A) Activin and TGFβ can stimulate biliary differentiation in fetal liver explants. E12.5 explants were cultured in the absence (Co) or in the presence of recombinant TGFβ-1 (TGFβ), activin A (ActA), or activin B (ActB) (in picograms per milliliter). Real-time RT–PCR quantification was performed for the biliary marker integrin_β_4 (ib4) or the hepatocyte markers cpsI and gys2, [(mRNA copy number)/(β-actin mRNA copy number)] × 103 (mean ± SEM). The expression of the biliary marker ggt4 was evaluated by semiquantitative RT–PCR, with tbp as control. (B) A local source of activin or TGFβ induces a gradient-wise expression of biliary cytokeratins in fetal liver explants. Beads (*) soaked into TGFβ-1, activin A, or activin B were implanted into E12.5 liver explants. After culture, sections were labeled for biliary cytokeratins. (C) TGFβ signaling is required for biliary differentiation. Wild-type pregnant mice were injected at E10.5 with anti-TGFβ-neutralizing antibodies or rabbit IgG as control. Sections collected at E14.5 were labeled for HNF-4α and biliary cytokeratins (CK) or laminin (Lam). (Arrowheads) Biliary basal lamina; (arrows) laminin produced by the mesenchyme surrounding the portal vein; (pv) portal vein. Bars, 100 μm.
Similar articles
- Control of hepatic differentiation by activin/TGFbeta signaling.
Clotman F, Lemaigre FP. Clotman F, et al. Cell Cycle. 2006 Jan;5(2):168-71. doi: 10.4161/cc.5.2.2341. Epub 2006 Jan 16. Cell Cycle. 2006. PMID: 16357531 - The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration.
Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, Lemaigre FP. Margagliotti S, et al. Dev Biol. 2007 Nov 15;311(2):579-89. doi: 10.1016/j.ydbio.2007.09.013. Epub 2007 Sep 16. Dev Biol. 2007. PMID: 17936262 - Transcription factor HNF-6/OC-1 inhibits the stimulation of the HNF-3alpha/Foxa1 gene by TGF-beta in mouse liver.
Plumb-Rudewiez N, Clotman F, Strick-Marchand H, Pierreux CE, Weiss MC, Rousseau GG, Lemaigre FP. Plumb-Rudewiez N, et al. Hepatology. 2004 Dec;40(6):1266-74. doi: 10.1002/hep.20459. Hepatology. 2004. PMID: 15562441 - [TGF-beta family (TGF-beta, activin, BMP)].
Koinuma D, Imamura T, Miyazono K. Koinuma D, et al. Nihon Rinsho. 2005 Apr;63 Suppl 4:215-9. Nihon Rinsho. 2005. PMID: 15861659 Review. Japanese. No abstract available. - [Identification of master regulators for induction of hepatocyte differentiation].
Suzuki A. Suzuki A. Seikagaku. 2012 Aug;84(8):675-9. Seikagaku. 2012. PMID: 23012878 Review. Japanese. No abstract available.
Cited by
- Hepatocyte polarity.
Treyer A, Müsch A. Treyer A, et al. Compr Physiol. 2013 Jan;3(1):243-87. doi: 10.1002/cphy.c120009. Compr Physiol. 2013. PMID: 23720287 Free PMC article. Review. - Lhx4 surpasses its paralog Lhx3 in promoting the differentiation of spinal V2a interneurons.
Renaux E, Baudouin C, Marchese D, Clovis Y, Lee SK, Gofflot F, Rezsohazy R, Clotman F. Renaux E, et al. Cell Mol Life Sci. 2024 Jul 6;81(1):286. doi: 10.1007/s00018-024-05316-x. Cell Mol Life Sci. 2024. PMID: 38970652 Free PMC article. - Interspecies Organogenesis for Human Transplantation.
Crane AT, Aravalli RN, Asakura A, Grande AW, Krishna VD, Carlson DF, Cheeran MC, Danczyk G, Dutton JR, Hackett PB, Hu WS, Li L, Lu WC, Miller ZD, O'Brien TD, Panoskaltsis-Mortari A, Parr AM, Pearce C, Ruiz-Estevez M, Shiao M, Sipe CJ, Toman NG, Voth J, Xie H, Steer CJ, Low WC. Crane AT, et al. Cell Transplant. 2019 Sep-Oct;28(9-10):1091-1105. doi: 10.1177/0963689719845351. Epub 2019 Aug 19. Cell Transplant. 2019. PMID: 31426664 Free PMC article. Review. - Optimal Hypoxia Regulates Human iPSC-Derived Liver Bud Differentiation through Intercellular TGFB Signaling.
Ayabe H, Anada T, Kamoya T, Sato T, Kimura M, Yoshizawa E, Kikuchi S, Ueno Y, Sekine K, Camp JG, Treutlein B, Ferguson A, Suzuki O, Takebe T, Taniguchi H. Ayabe H, et al. Stem Cell Reports. 2018 Aug 14;11(2):306-316. doi: 10.1016/j.stemcr.2018.06.015. Epub 2018 Jul 19. Stem Cell Reports. 2018. PMID: 30033085 Free PMC article. - Mechanisms of hepatocyte growth factor-mediated and epidermal growth factor-mediated signaling in transdifferentiation of rat hepatocytes to biliary epithelium.
Limaye PB, Bowen WC, Orr AV, Luo J, Tseng GC, Michalopoulos GK. Limaye PB, et al. Hepatology. 2008 May;47(5):1702-13. doi: 10.1002/hep.22221. Hepatology. 2008. PMID: 18398918 Free PMC article.
References
- Annes J.P., Munger, J.S., and Rifkin, D.B. 2003. Making sense of latent TGFβ activation. J. Cell Sci. 116: 217–224. - PubMed
- Arandjelovic S., Freed, T.A., and Gonias, S.L. 2003. Growth factor-binding sequence in human α2-macroglobulin targets the receptor-binding site in transforming growth factor-β. Biochemistry 42: 6121–6127. - PubMed
- Clotman F., Lannoy, V.J., Reber, M., Cereghini, S., Cassiman, D., Jacquemin, P., Roskams, T., Rousseau, G.G., and Lemaigre, F.P. 2002. The onecut transcription factor Hnf6 is required for normal development of the biliary tract. Development 129: 1819–1828. - PubMed
- Desmet V.J. 1992. Congenital diseases of intrahepatic bile ducts: Variations on the theme `ductal plate malformation.' Hepatology 16: 1069–1083. - PubMed
- de Sousa Lopes S.M., Carvalho, R.L., van den Driesche, S., Goumans, M.J., ten Dijke, P., and Mummery, C.L. 2003. Distribution of phosphorylated Smad2 identifies target tissues of TGF β ligands in mouse development. Gene Expr. Patterns 3: 355–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases