The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes - PubMed (original) (raw)
The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes
Mikael Sjölinder et al. Genes Dev. 2005.
Abstract
In the dipteran Chironomus tentans, actin binds to hrp65, a nuclear protein associated with mRNP complexes. Disruption of the actin-hrp65 interaction in vivo by the competing peptide 65-2CTS reduces transcription drastically, which suggests that the actin-hrp65 interaction is required for transcription. We show that the inhibitory effect of the 65-2CTS peptide on transcription is counteracted by trichostatin A, a drug that inhibits histone deacetylation. We also show that actin and hrp65 are associated in vivo with p2D10, an evolutionarily conserved protein with histone acetyltransferase activity that acts on histone H3. p2D10 is recruited to class II genes in a transcription-dependent manner. We show, using the Balbiani ring genes of C. tentans as a model system, that p2D10 is cotranscriptionally associated with the growing pre-mRNA. We also show that experimental disruption of the actin-hrp65 interaction by the 65-2CTS peptide in vivo results in the release of p2D10 from the transcribed genes, reduced histone H3 acetylation, and a lower level of transcription activity. Furthermore, antibodies against p2D10 inhibit run-on elongation. Our results suggest that actin, hrp65, and p2D10 are parts of a positive feedback mechanism that contributes to maintaining the active transcription state of a gene by recruiting HATs at the RNA level.
Figures
Figure 1.
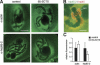
Association of p2D10 with class II genes. Polytene chromosome squashes from untreated larvae and from larvae grown in the presence of galactose were immunostained with the anti-p2D10 mAb 1F2. In untreated chromosomes, the antibody stained the active BR1, BR2, and BR3 gene loci on chromosome IV, and it stained other loci in the polytene chromosomes (arrows). This pattern shows that p2D10 is located at transcriptionally active class II genes. Feeding the larvae with galactose induced the expression of the BR6 gene on chromosome III and the recruitment of p2D10 to the activated BR6 locus. The galactose treatment also reduced the transcriptional activity at the BR2 locus, and this was accompanied by a decrease in the p2D10 signal at this site (cf. the staining at BR2 in untreated and galactose). The micrographs were taken under bright-field conditions. (BR) Balbiani ring; (NU) nucleolus. The scale bar represents ∼10 μm.
Figure 2.
p2D10 is associated with growing BR pre-mRNP particles. (A) Schematic representation of the active BR transcription unit illustrating the distinct morphology of the growing pre-mRNPs in the proximal (p), middle (m), and distal (d) portions of the BR gene. (B) Immuno-EM analysis of p2D10 distribution in the active BR transcription unit. Thin cryosections of aldehyde-fixed salivary glands were immunolabeled with anti-p2D10 mAbs, either 2D10 or 1F2, and detected with a secondary antibody conjugated to colloidal gold markers. The left panel shows a section of a salivary gland cell nucleus immunostained with anti-p2D10 antibodies showing the different portions of a BR gene as visualized in the EM. The right panel shows a schematic interpretation of the micrograph. The bar represents 200 nm. (C) The histogram shows the distribution of p2D10 along the active BR gene obtained with two independent mAbs against p2D10, mAb 2D10 and mAb 1F2. For each antibody, the numbers of immuno-gold markers located in each portion of the BR gene were counted, and the distribution of labeling was expressed as percentage of immuno-gold markers in each portion of the gene. The number of gold markers analyzed was n = 402 for mAb 2D10 and n = 1730 for mAb 1F2. The distribution of hrp65-2, based on data from Percipalle et al. (2003), is also presented for comparison. (D) Four examples of immunolabeling in growing BR pre-mRNP particles at higher magnification. Note the association of immuno-gold markers with the growing BR pre-mRNP particles. Schematic interpretations of the images are provided under each micrograph. The scale bar represents 100 nm. (E) The association of p2D10 with the genes is RNA-dependent. Salivary glands were permeabilized and digested with RNase A before immunofluorescent staining with mAb 1F2 against p2D10. The figure shows confocal sections through the nuclei of salivary gland cells treated with (right panel) and without (control, left panel) RNase A. (CYT) Cytoplasm; (NUC) nucleus. The magnification bar represents 25 μm.
Figure 3.
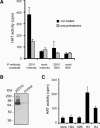
The histone acetyltransferase activity of p2D10. (A) p2D10 was immunoprecipitated from nuclear extracts and HAT activity was determined in solution using a mixture of core histones as substrate. Immunoprecipitations were performed according to standard protocols under native conditions (black bars) or using urea-treated extracts to dissociate protein complexes (gray bars). Control experiments were performed without specific antibody (mock) or with an alternative substrate, BSA, as indicated. (B) Lysates of bacteria expressing recombinant p2D10 or empty vector (control) were subjected to in-gel HAT assays. (C) Histone specificity was determined by immunoprecipitating p2D10 and determining HAT activity in solution using individual histones as substrate.
Figure 4.
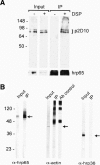
Interaction of p2D10 with actin and hrp65 in vivo. (A) p2D10 binds directly to hrp65. C. tentans tissue culture cells were treated with DSP in vivo, and nuclear extracts were prepared. Extracts were treated with urea to dissociate non-cross-linked proteins and subjected to immunoprecipitation using mAb 4E9 against hrp65. The immunoprecipitated material (IP) and the starting material (Input) were analyzed by Western blotting with anti-p2D10 antibodies. DSP cross-linking was reversed by the reducing SDS-PAGE sample buffer. (B) Actin and hrp65 are coimmunoprecipitated by anti-p2D10 antibodies. p2D10 was immunoprecipitated from nuclear extracts prepared from C. tentans tissue culture cells using a peptide-specific antibody directed against p2D10. The starting material (Input) and precipitated material (IP) were analyzed by Western blotting with the indicated antibodies. The coimmunoprecipitated proteins are indicated by arrows. hrp36, an hnRNP protein abundant in the Input but not detected in the IP fraction, was assayed in parallel as a negative control to assess the specificity of the immunoprecipitation. In the detection of actin, the anti-rabbit antibodies used in the second step of the Western blot reacted against the primary anti-p2D10 antibody used in the immunoprecipitation, which results in several intense bands, as shown by a control immunoprecipitation carried out with the same antibodies without protein extract.
Figure 5.
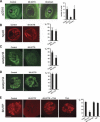
Effects of the 65-2CTS peptide on p2D10 location, histone acetylation, and transcription. Each picture shows a confocal section through the nucleus of a salivary gland cell immunostained as indicated. (A) Salivary glands dissected from fourth instar larvae were incubated for 30 min in culture medium with or without (Control) peptides, as indicated, and subsequently immunostained with mAb 1F2 against p2D10. The number of nuclei per gland showing intense chromosomal staining was recorded, and the results of the quantification are summarized in the histogram. Four glands from two independent experiments were analyzed in each case. Treatment of the glands with 65-2CTS peptide caused a specific reduction in the association of p2D10 with the chromosomes. (Nu) Nucleolus. (B) Control and treated glands were processed as above and immunostained with mAb 2E4 against hrp45. The association of hrp45 with the chromosomes is not affected by treatment with the 65-2CTS peptide. Eight glands from two independent experiments were analyzed in each case. (C,D) Control and treated glands were processed as above and stained with rabbit antibodies against either acetylated histone H3 (C) or acetylated histone H4 (D). The histograms show the percentage of nuclei per gland with intense nuclear staining. Twelve independent glands were analyzed in each case. Note that treatment of the glands with the 65-2CTS peptide resulted in specific reduction of histone H3 acetylation in vivo. (E) Salivary glands were dissected and incubated for 30 min with or without 65-2CTS peptide and/or trichostatin A (TSA) as indicated, and the transcriptional activity of the glands was sub-sequently assayed by BrUTP incorporation. The histogram shows the percentage of nuclei per gland with intense BrUTP staining. The number of glands analyzed in each case was n = 9, 8, 9, and 4 for control, 65-2CTS, 65-2CTS + TSA, and TSA, respectively. In all cases, error bars indicate standard deviations, and magnification bars represent 25 μm. Results from two independent experiments were pooled.
Figure 6.
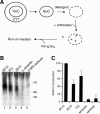
Specific inhibition of run-on transcription by anti-p2D10 antibodies. (A) Schematic depiction of the run-on assay to test global effects of specific antibodies on transcription elongation. (B) Permeabilized nuclei were prepared from cultured C. tentans cells, incubated with or without antibodies as indicated, and subjected to transcription run-on analysis. mAbs 1F2 (lane 2) and 2D10 (lane 3) are against p2D10, and mAb 6C10 (lane 1) is against an unrelated chromosomal protein of 35 kDa. In lane 4, the run-on reaction was performed in the presence of 3 μg/mL α-amanitin. The run-on reactions in lanes 1_–_4 were performed in the presence of 0.015% sarkosyl, whereas the reaction in lane 5 is a negative control with 0.08% sarkosyl. The RNA elongated in the run-on was resolved in urea–polyacrylamide gels, and the levels of ribonucleotide incorporation into RNA were measured by phosphorimaging in the 200–700-nt range. (C) The histogram shows the relative levels of ribonucleotide incorporation into RNA determined by PhosphorImager analysis. Values indicate mean ± standard deviation of three independent experiments.
Figure 7.
The association of hrp65-2 with the genes is disrupted by the 65-2CTS peptide. Each picture shows a confocal section through the nucleus of a salivary gland cell immunostained as indicated. (A) Salivary glands dissected from fourth instar larvae were incubated for 30 min in culture medium with or without (control) peptides, as indicated, and subsequently immunostained with antibodies against either actin or hrp65-2. In control glands, both proteins showed intense nucleoplasmic accumulation, and positively stained bands in the chromosomes. Treatment of the glands with the 65-2CTS peptide did not change the location of actin but reduced the levels of chromosomal hrp65-2. (B) Salivary glands treated as above were double-stained with mAb anti-hrp45, used as a marker for BR puffs, and either anti-actin or anti-hrp65-2 antibodies. The micrograph shows a control chromosome stained for hrp65-2 (green) and hrp45 (red). Possible changes in the association of actin and hrp65-2 with transcriptionally active loci were analyzed by selecting BR puffs on the basis of the hrp45 staining (red), and measuring the intensity of labeling in the BR puffs relative to hrp45, as described in the Results. (C) The results of the quantitative analysis. The histogram shows the levels of fluorescence for actin and hrp65-2, relative to hrp45, in BR puffs from control and treated glands, as indicated. Values indicate mean ± standard deviation. n = 16 and 19 for actin control and treated, respectively, and n = 20 and 25 for hrp65-2 control and treated, respectively. Results from two independent experiments were pooled. The scale bars represent ∼20 μm in A and 10 μm in B.
Figure 8.
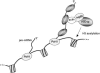
A model for the cotranscriptional recruitment of p2D10 to the growing pre-mRNP. Actin, hrp65, and p2D10 become incorporated into the growing pre-mRNP cotranscriptionally. This serves to localize p2D10 close to its substrate, histone H3. We do not know whether p2D10 can reach and acetylate H3 while bound to the pre-mRNP, or whether the HAT activity of p2D10 is dependent on high on–off rates. The proposed order of assembly of the proteins on the pre-mRNA is based on reported protein–protein interactions and on the effect of the 65-2CTS peptide on the association of actin, hrp65, and p2D10 with the chromosomes. Although binding of the actin–hrp65–p2D10 complex to the growing RNA could be mediated by the RNA-binding domains of hrp65, we favor the hypothesis that actin mediates such association by binding to hnRNP proteins.
Similar articles
- An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II.
Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, Visa N. Percipalle P, et al. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6475-80. doi: 10.1073/pnas.1131933100. Epub 2003 May 12. Proc Natl Acad Sci U S A. 2003. PMID: 12743363 Free PMC article. - Evidence for a posttranscriptional role of a TFIIICalpha-like protein in Chironomus tentans.
Sabri N, Farrants AK, Hellman U, Visa N. Sabri N, et al. Mol Biol Cell. 2002 May;13(5):1765-77. doi: 10.1091/mbc.01-09-0436. Mol Biol Cell. 2002. PMID: 12006668 Free PMC article. - The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm.
Zhao J, Jin SB, Björkroth B, Wieslander L, Daneholt B. Zhao J, et al. EMBO J. 2002 Mar 1;21(5):1177-87. doi: 10.1093/emboj/21.5.1177. EMBO J. 2002. PMID: 11867546 Free PMC article. - Molecular functions of nuclear actin in transcription.
Percipalle P, Visa N. Percipalle P, et al. J Cell Biol. 2006 Mar 27;172(7):967-71. doi: 10.1083/jcb.200512083. Epub 2006 Mar 20. J Cell Biol. 2006. PMID: 16549500 Free PMC article. Review. - Retinoid receptors in health and disease: co-regulators and the chromatin connection.
Minucci S, Pelicci PG. Minucci S, et al. Semin Cell Dev Biol. 1999 Apr;10(2):215-25. doi: 10.1006/scdb.1999.0303. Semin Cell Dev Biol. 1999. PMID: 10441075 Review.
Cited by
- Nuclear translocation of beta-actin is involved in transcriptional regulation during macrophage differentiation of HL-60 cells.
Xu YZ, Thuraisingam T, Morais DA, Rola-Pleszczynski M, Radzioch D. Xu YZ, et al. Mol Biol Cell. 2010 Mar 1;21(5):811-20. doi: 10.1091/mbc.e09-06-0534. Epub 2010 Jan 6. Mol Biol Cell. 2010. PMID: 20053683 Free PMC article. - Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing.
Saldi T, Cortazar MA, Sheridan RM, Bentley DL. Saldi T, et al. J Mol Biol. 2016 Jun 19;428(12):2623-2635. doi: 10.1016/j.jmb.2016.04.017. Epub 2016 Apr 20. J Mol Biol. 2016. PMID: 27107644 Free PMC article. Review. - Nuclear actin and myosins: life without filaments.
de Lanerolle P, Serebryannyy L. de Lanerolle P, et al. Nat Cell Biol. 2011 Nov 2;13(11):1282-8. doi: 10.1038/ncb2364. Nat Cell Biol. 2011. PMID: 22048410 Review. - The long journey of actin and actin-associated proteins from genes to polysomes.
Percipalle P. Percipalle P. Cell Mol Life Sci. 2009 Jul;66(13):2151-65. doi: 10.1007/s00018-009-0012-8. Epub 2009 Mar 20. Cell Mol Life Sci. 2009. PMID: 19300907 Free PMC article. Review. - Integration of mRNP formation and export.
Björk P, Wieslander L. Björk P, et al. Cell Mol Life Sci. 2017 Aug;74(16):2875-2897. doi: 10.1007/s00018-017-2503-3. Epub 2017 Mar 17. Cell Mol Life Sci. 2017. PMID: 28314893 Free PMC article. Review.
References
- Alzhanova-Ericsson A.T., Sun, X., Visa, N., Kiseleva, E., Wurtz, T., and Daneholt, B. 1996. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes & Dev. 10: 2881–2893. - PubMed
- Beermann W. 1973. Directed changes in the pattern of Balbiani ring puffing in Chironomus: Effects of a sugar treatment. Chromosoma 41: 297–326. - PubMed
- Bettinger B.T., Gilbert, D.M., and Amberg, D.C. 2004. Actin up in the nucleus. Nat. Rev. Mol. Cell. Biol. 5: 410–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources