Ancestral antibiotic resistance in Mycobacterium tuberculosis - PubMed (original) (raw)
Comparative Study
. 2005 Aug 23;102(34):12200-5.
doi: 10.1073/pnas.0505446102. Epub 2005 Aug 15.
Liem Nguyen, John Gatfield, Kevin Visconti, Kien Nguyen, Dirk Schnappinger, Sabine Ehrt, Yang Liu, Leonid Heifets, Jean Pieters, Gary Schoolnik, Charles J Thompson
Affiliations
- PMID: 16103351
- PMCID: PMC1186028
- DOI: 10.1073/pnas.0505446102
Comparative Study
Ancestral antibiotic resistance in Mycobacterium tuberculosis
Rowan P Morris et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Chemotherapeutic options to treat tuberculosis are severely restricted by the intrinsic resistance of Mycobacterium tuberculosis to the majority of clinically applied antibiotics. Such resistance is partially provided by the low permeability of their unique cell envelope. Here we describe a complementary system that coordinates resistance to drugs that have penetrated the envelope, allowing mycobacteria to tolerate diverse classes of antibiotics that inhibit cytoplasmic targets. This system depends on whiB7, a gene that pathogenic Mycobacterium shares with Streptomyces, a phylogenetically related genus known as the source of diverse antibiotics. In M. tuberculosis, whiB7 is induced by subinhibitory concentrations of antibiotics (erythromycin, tetracycline, and streptomycin) and whiB7 null mutants (Streptomyces and Mycobacterium) are hypersusceptible to antibiotics in vitro. M. tuberculosis is also antibiotic sensitive within a monocyte model system. In addition to antibiotics, whiB7 is induced by exposure to fatty acids that pathogenic Mycobacterium species may accumulate internally or encounter within eukaryotic hosts during infection. Gene expression profiling analyses demonstrate that whiB7 transcription determines drug resistance by activating expression of a regulon including genes involved in ribosomal protection and antibiotic efflux. Components of the whiB7 system may serve as attractive targets for the identification of inhibitors that render M. tuberculosis or multidrug-resistant derivatives more antibiotic-sensitive.
Figures
Fig. 1.
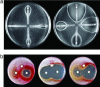
Identification of whiB7, a gene providing intrinsic antibiotic resistance in Streptomyces. (a) Antibiotic susceptibility of the wild-type S. lividans strain (Left) and the spontaneous whiB7 mutant RM1 (Right). Etest strips were applied to seeded spores, and minimal inhibition concentration values were read from the scale (μg/ml) at the point of intersection between inhibition ellipse edge and the strip. Upper, erythromycin; right, tetracycline; lower, rifampicin; left, quinupristin/dalfopristin. (b) S. lividans RM1 was engineered to allow thiostrepton-inducible expression of whiB7 by using the expression plasmid pIJ8600. Seeded spores of S. lividans wild-type/pIJ8600 (Left), the whiB7 mutant RM1/pIJ8600 (Center), or RM1/pIJ8600::whiB7 (Right) were exposed to radial gradients by discs containing 100 μg of oleandomycin (left), chlorotetracycline (right), or thiostrepton (top). The high frequency of suppressor colonies in the RM1/pIJ8600::whiB7 culture are presumed to be promoter-up mutants that allow unregulated whiB7 expression. Tetracycline induced synthesis of a red pigment (likely to be the antibiotic undecylprodigiosin) in both wild-type S. lividans and the whiB7 mutant.
Fig. 2.
The orthologous whiB7 loci of Streptomyces and Mycobacterium. (a) Alignment of the WhiB7 proteins from both Mycobacterium tuberculosis (WhiB7-tub) and leprae (WhiB7-lep) with the Streptomyces WhiB7 and the prototypic family member WhiB from Streptomyces coelicolor. Four absolutely conserved cysteine residues and a tryptophan-containing/glycine-rich motif are conserved throughout the WhiB family (circled). An A/T-Hook DNA binding consensus sequence is found only in WhiB7 paralogs. ∼, N-terminal sequence not shown. (b) Gene organization of the whiB7 genomic region. Shaded block arrows represent conserved ORFs.
Fig. 3.
Identification of antibiotic resistance genes as parts of the M. tuberculosis whiB7 regulon. Significantly altered gene expression ratios from all experiments were averaged, log2 transformed, and clustered according to the displayed color code. Red and blue indicate higher and lower gene expression, respectively, in experimental samples, in comparison to the reference. Black represents no difference. (a) Genes of the whiB7 regulon. Rv and ORF numbers designate annotated M. tuberculosis genes, and arrows indicate contiguous genes. (b) Treatment (15 min) of M. tuberculosis 1254 with 0.5, 1.0, 5, 25 and 100 μg/ml of erythromycin, streptomycin, and tetracycline. (c) Extended antibiotic treatment for 2 and 24 h (1 μg/ml). (d) M. tuberculosis H37Rv was used as reference and compared to a whiB7 null mutant (WhiB7 KO) and a H37Rv strain engineered to overexpress whiB7 (WhiB7 OV). Cells were assayed at an optical density of 0.4 at 600 nm. (e) whiB7 induction by fatty acids. Quantitative RT-PCR-determined induction factors of whiB7 expression. Primary y axis: M. tuberculosis grown in MDG fed 50 μM palmitic acid (purple) or its unsaturated form, oleic acid (gray). Secondary y axis: induction to prolonged exposure of tetracycline (connected dots) (≈2 μM, 1 μg/ml).
Fig. 4.
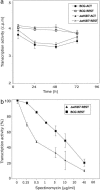
The phenotype of the whiB7 mutation in J774, a monocyte-derived cell line. Mycobacterial RNA transcription, an indicator of survival, was assayed by incorporation of tritiated uracil. Shown are mean values (±SD) of six to eight determinations. Resting (REST) or activated (ACT) J774 monocytes growing in microtiter plates were exposed to M. bovis bacillus Calmette-Guérin and the corresponding whiB7 mutant. (a) RNA synthesis of internalized WT and whiB7 mutant cells decreased at the same rate in resting or activated monocytes, presumably because of bactericidal activity of the monocytes. In contrast, growth did occur in the control experiment where the bacteria were cultured in the same medium without monocytes (data not shown). (b) For antibiotic susceptibility testing, infected monocytes were incubated with medium containing the indicated concentrations of spectinomycin; % transcriptional activity is the percentage of incorporation rates in spectinomycin-treated vs. untreated bacteria. The same results were obtained in infected monocytes treated with amikacin after infection to confirm that incorporation rates reflected internalized mycobacteria (data not shown).
Similar articles
- The WblC/WhiB7 Transcription Factor Controls Intrinsic Resistance to Translation-Targeting Antibiotics by Altering Ribosome Composition.
Lee JH, Yoo JS, Kim Y, Kim JS, Lee EJ, Roe JH. Lee JH, et al. mBio. 2020 Apr 14;11(2):e00625-20. doi: 10.1128/mBio.00625-20. mBio. 2020. PMID: 32291305 Free PMC article. - WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis.
Burian J, Ramón-García S, Howes CG, Thompson CJ. Burian J, et al. Expert Rev Anti Infect Ther. 2012 Sep;10(9):1037-47. doi: 10.1586/eri.12.90. Expert Rev Anti Infect Ther. 2012. PMID: 23106278 Review. - Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Lilic M, Darst SA, Campbell EA. Lilic M, et al. Mol Cell. 2021 Jul 15;81(14):2875-2886.e5. doi: 10.1016/j.molcel.2021.05.017. Epub 2021 Jun 24. Mol Cell. 2021. PMID: 34171296 Free PMC article. - uORF-mediated riboregulation controls transcription of whiB7/wblC antibiotic resistance gene.
Lee JH, Lee EJ, Roe JH. Lee JH, et al. Mol Microbiol. 2022 Jan;117(1):179-192. doi: 10.1111/mmi.14834. Epub 2021 Nov 2. Mol Microbiol. 2022. PMID: 34687261 - The function and regulatory network of WhiB and WhiB-like protein from comparative genomics and systems biology perspectives.
Zheng F, Long Q, Xie J. Zheng F, et al. Cell Biochem Biophys. 2012 Jun;63(2):103-8. doi: 10.1007/s12013-012-9348-z. Cell Biochem Biophys. 2012. PMID: 22388511 Review.
Cited by
- Bacterial iron-sulfur regulatory proteins as biological sensor-switches.
Crack JC, Green J, Hutchings MI, Thomson AJ, Le Brun NE. Crack JC, et al. Antioxid Redox Signal. 2012 Nov 1;17(9):1215-31. doi: 10.1089/ars.2012.4511. Epub 2012 Mar 6. Antioxid Redox Signal. 2012. PMID: 22239203 Free PMC article. Review. - Unraveling the mechanisms of intrinsic drug resistance in Mycobacterium tuberculosis.
Poulton NC, Rock JM. Poulton NC, et al. Front Cell Infect Microbiol. 2022 Oct 17;12:997283. doi: 10.3389/fcimb.2022.997283. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36325467 Free PMC article. Review. - Molecular mechanisms of underlying genetic factors and associated mutations for drug resistance in Mycobacterium tuberculosis.
Swain SS, Sharma D, Hussain T, Pati S. Swain SS, et al. Emerg Microbes Infect. 2020 Dec;9(1):1651-1663. doi: 10.1080/22221751.2020.1785334. Emerg Microbes Infect. 2020. PMID: 32573374 Free PMC article. Review. - Incomplete transcripts dominate the Mycobacterium tuberculosis transcriptome.
Ju X, Li S, Froom R, Wang L, Lilic M, Delbeau M, Campbell EA, Rock JM, Liu S. Ju X, et al. Nature. 2024 Mar;627(8003):424-430. doi: 10.1038/s41586-024-07105-9. Epub 2024 Feb 28. Nature. 2024. PMID: 38418874 Free PMC article. - Genome plasticity of BCG and impact on vaccine efficacy.
Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, Inwald JK, Golby P, Garcia JN, Hewinson RG, Behr MA, Quail MA, Churcher C, Barrell BG, Parkhill J, Cole ST. Brosch R, et al. Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5596-601. doi: 10.1073/pnas.0700869104. Epub 2007 Mar 19. Proc Natl Acad Sci U S A. 2007. PMID: 17372194 Free PMC article.
References
- Dye, C., Scheele, S., Dolin, P., Pathania, V. & Raviglione, M. C. (1999) J. Am. Med. Assoc. 282, 677-686. - PubMed
- McKinney, J. D. (2000) Nat. Med. 6, 1330-1333. - PubMed
- Parrish, N. M., Dick, J. D. & Bishai, W. R. (1998) Trends Microbiol. 6, 107-112. - PubMed
- Jarlier, V. & Nikaido, H. (1994) FEMS Microbiol. Lett. 123, 11-18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases