Mimivirus relatives in the Sargasso sea - PubMed (original) (raw)
Mimivirus relatives in the Sargasso sea
Elodie Ghedin et al. Virol J. 2005.
Abstract
The discovery and genome analysis of Acanthamoeba polyphaga Mimivirus, the largest known DNA virus, challenged much of the accepted dogma regarding viruses. Its particle size (>400 nm), genome length (1.2 million bp) and huge gene repertoire (911 protein coding genes) all contribute to blur the established boundaries between viruses and the smallest parasitic cellular organisms. Phylogenetic analyses also suggested that the Mimivirus lineage could have emerged prior to the individualization of cellular organisms from the three established domains, triggering a debate that can only be resolved by generating and analyzing more data. The next step is then to seek some evidence that Mimivirus is not the only representative of its kind and determine where to look for new Mimiviridae. An exhaustive similarity search of all Mimivirus predicted proteins against all publicly available sequences identified many of their closest homologues among the Sargasso Sea environmental sequences. Subsequent phylogenetic analyses suggested that unknown large viruses evolutionarily closer to Mimivirus than to any presently characterized species exist in abundance in the Sargasso Sea. Their isolation and genome sequencing could prove invaluable in understanding the origin and diversity of large DNA viruses, and shed some light on the role they eventually played in the emergence of eukaryotes.
Figures
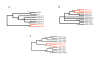
Figure 1
Phylogenetic evidence of uncharacterized Mimivirus relatives. (a) Neighbor-joining (NJ) clustering (see Materials and Methods) of Mimivirus R449 ORF with its best matching (≈35% identical residues) environmental homologues (noted Sargasso1 to Sargasso6 according to their decreasing similarity) and closest viral orthologues (28% identical). (b) NJ clustering of Mimivirus R429 ORF with its best matching (≈50% identical) environmental homologues (noted Sargasso1 to Sargasso5) and closest viral orthologues (35% identical). (c) NJ clustering of Mimivirus putative virion packaging ATPase L437 with its best matching (≈45% identity) environmental homologues (Sargasso1 and Sargasso2) and closest viral orthologues (34% identical). Abbreviations: Phyco: Phycodnavirus; PBCV: Paramecium bursaria chlorella virus 1; EsV: Ectocarpus siliculosus virus; FsV: Feldmannia sp. virus; HaV: Heterosigma akashiwo virus; Irido: Iridovirus; LCDV: Lymphocystis disease virus 1; Frog: Frog virus 3; Amby: Ambystoma tigrinum stebbensi virus; Rana: Rana tigrina ranavirus; Chilo: Chilo iridescent virus. Bootstrap values larger than 50% are shown. Branches with lower values were condensed.
Figure 2
Organization of four Mimivirus ORF best matching homologues in a 4.5 kb environmental sequence scaffold (approximately to scale). The three colinear Mimivirus homologues are in green. Unmatched ORF extremities are indicated by dots. The two diagonal lines indicate where the two contigs are joined on the scaffold.
Figure 3
Partial 3-way alignment (N-terminus region) of Mimivirus capsid protein (R441) with it best matching homologues in the NR and Environmental sequence databases. The Mimivirus R441 protein shares 83/229 (36.2%) identical residues (colored in red or blue) with the major capsid protein Vp49 of Chlorella virus CVG-1 and 111/229 (48.5%) identical residues (indicated in red or green) with the N-terminus of a capsid protein from an unknown large virus sampled from the Sargasso Sea (Accession: EAD00518). On the other hand, the CVG-1 Vp49 and the Sargasso Sea sequence share 44.5% identical residues. By comparison, the CVG-1 Vp49 protein share 72% of identical residue with PBCV-1 Vp54, its best matching homologue among known phycodnaviruses.
References
- Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol. 2001;52:419–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources