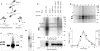Endonucleolytic processing of covalent protein-linked DNA double-strand breaks - PubMed (original) (raw)
Endonucleolytic processing of covalent protein-linked DNA double-strand breaks
Matthew J Neale et al. Nature. 2005.
Abstract
DNA double-strand breaks (DSBs) with protein covalently attached to 5' strand termini are formed by Spo11 to initiate meiotic recombination. The Spo11 protein must be removed for the DSB to be repaired, but the mechanism for removal is unclear. Here we show that meiotic DSBs in budding yeast are processed by endonucleolytic cleavage that releases Spo11 attached to an oligonucleotide with a free 3'-OH. Two discrete Spo11-oligonucleotide complexes were found in equal amounts, differing with respect to the length of the bound DNA. We propose that these forms arise from different spacings of strand cleavages flanking the DSB, with every DSB processed asymmetrically. Thus, the ends of a single DSB may be biochemically distinct at or before the initial processing step-much earlier than previously thought. SPO11-oligonucleotide complexes were identified in extracts of mouse testis, indicating that this mechanism is evolutionarily conserved. Oligonucleotide-topoisomerase II complexes were also present in extracts of vegetative yeast, although not subject to the same genetic control as for generating Spo11-oligonucleotide complexes. Our findings suggest a general mechanism for repair of protein-linked DSBs.
Conflict of interest statement
Competing interests statement: The authors declare that they have no competing financial interests.
Figures
Figure 1
Endonucleolytic processing of covalent Spo11-DSB complexes. a, Alternative mechanisms for Spo11 release. b, Detection of Spo11-oligonucleotide complexes. Immunoprecipitates from the indicated genotypes with or without antibody were labelled with TdT. Upper panel, autoradiograph; lower panel, anti-HA western. c, Relative mobilities of free and oligonucleotide-associated Spo11-HA. Panel 1, autoradiograph; panels 2–3, low and high exposures of an anti-HA western; panel 4, re-exposure of the blot to film after partial fading of the chemiluminescent signal. d, Sizes of Spo11-associated oligonucleotides from the upper (U) and lower (L) SDS-PAGE bands. Size standards are indicated. e, Time course of appearance of Spo11-oligonucleotide complexes during meiosis. f, Quantification of upper (⋄) and lower (Δ) labelled Spo11 species from e, and the DSB frequency at the HIS4LEU2 recombination hotspot measured in the same culture (▪). Each point is a single measurement. Asterisks, non-specific labelling; closed arrows, Spo11-specific labelled species; open arrows, free Spo11-HA.
Figure 2
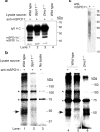
SPO11-oligonucleotide complexes in mouse meiosis. a, Testis extracts immunoprecipitated with or without anti-mSPO11 were probed by western blotting with the same antibody. Each sample represents extract from an equivalent number of testes. IgG H.C., immunoglobulin heavy chain. b, mSPO11-oligonucleotide complexes. mSPO11 was immunoprecipitated from testis extracts and labelled with TdT. Samples were from two testes (lanes 1–2) or six testes (lanes 4–6). Arrows, mSPO11-specific species; asterisks, non-specific bands. c, Sizes of mSPO11-associated oligonucleotides after gel purification and protease digestion. A mock immunoprecipitation lacking the anti-mSPO11 antibody was processed in parallel.
Figure 3
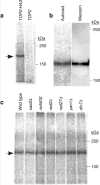
Topoisomerase II-oligonucleotide complexes in non-meiotic yeast cells. a, Extracts were prepared from vegetatively growing yeast strains carrying topoisomerase II with (TOP2-HA3) or without (TOP2) an epitope tag. Samples were immunoprecipitated with anti-HA antibody and labelled with TdT. b, Comparison of electrophoretic mobilities of free and oligonucleotide-associated Top2-HA3. Immunoprecipitated Top2-HA3 was labelled with TdT, separated on 5% SDS-PAGE, and transferred to PVDF membrane. Autoradiograph and anti-HA western of the membrane are shown. c, Formation of Top2-oligonucleotide complexes in nuclease-defective mutants. Arrow, Top2-HA3-specific band.
Figure 4
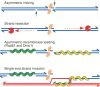
Asymmetric steps in meiotic recombination. A Spo11 dimer (orange elipses) creates a DSB which is processed by asymmetrically spaced nicks. Exonucleolytic resection initiates at these nicks, yielding single-stranded gaps. Rad51 and Dmc1 recombinases (green and yellow, without specifying which) form nucleoprotein filaments on opposite sides of the DSB,. Asymmetric strand invasion yields a stable strand exchange intermediate. Spo11-oligonucleotide complexes, stabilized by base pairing and protein-protein interactions, are proposed to remain associated with DSB ends until a step at or subsequent to strand exchange. See text for further details.
Similar articles
- Exploring the removal of Spo11 and topoisomerases from DNA breaks in S. cerevisiae by human Tyrosyl DNA Phosphodiesterase 2.
Johnson D, Allison RM, Cannavo E, Cejka P, Harper JA, Neale MJ. Johnson D, et al. DNA Repair (Amst). 2024 Oct;142:103757. doi: 10.1016/j.dnarep.2024.103757. Epub 2024 Aug 31. DNA Repair (Amst). 2024. PMID: 39236418 - Modulating and targeting meiotic double-strand breaks in Saccharomyces cerevisiae.
Nicolas A. Nicolas A. Methods Mol Biol. 2009;557:27-33. doi: 10.1007/978-1-59745-527-5_3. Methods Mol Biol. 2009. PMID: 19799174 Review. - ATM controls meiotic double-strand-break formation.
Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. Lange J, et al. Nature. 2011 Oct 16;479(7372):237-40. doi: 10.1038/nature10508. Nature. 2011. PMID: 22002603 Free PMC article. - Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases.
Manfrini N, Guerini I, Citterio A, Lucchini G, Longhese MP. Manfrini N, et al. J Biol Chem. 2010 Apr 9;285(15):11628-37. doi: 10.1074/jbc.M110.104083. Epub 2010 Feb 11. J Biol Chem. 2010. PMID: 20150422 Free PMC article. - End-labeling and analysis of Spo11-oligonucleotide complexes in Saccharomyces cerevisiae.
Neale MJ, Keeney S. Neale MJ, et al. Methods Mol Biol. 2009;557:183-95. doi: 10.1007/978-1-59745-527-5_12. Methods Mol Biol. 2009. PMID: 19799183 Free PMC article. Review.
Cited by
- Coupling end resection with the checkpoint response at DNA double-strand breaks.
Villa M, Cassani C, Gobbini E, Bonetti D, Longhese MP. Villa M, et al. Cell Mol Life Sci. 2016 Oct;73(19):3655-63. doi: 10.1007/s00018-016-2262-6. Epub 2016 May 3. Cell Mol Life Sci. 2016. PMID: 27141941 Free PMC article. Review. - Control of Meiotic Crossovers: From Double-Strand Break Formation to Designation.
Gray S, Cohen PE. Gray S, et al. Annu Rev Genet. 2016 Nov 23;50:175-210. doi: 10.1146/annurev-genet-120215-035111. Epub 2016 Sep 14. Annu Rev Genet. 2016. PMID: 27648641 Free PMC article. Review. - A covalent protein-DNA 5'-product adduct is generated following AP lyase activity of human ALKBH1 (AlkB homologue 1).
Müller TA, Andrzejak MM, Hausinger RP. Müller TA, et al. Biochem J. 2013 Jun 15;452(3):509-18. doi: 10.1042/BJ20121908. Biochem J. 2013. PMID: 23577621 Free PMC article. - Competing roles of DNA end resection and non-homologous end joining functions in the repair of replication-born double-strand breaks by sister-chromatid recombination.
Muñoz-Galván S, López-Saavedra A, Jackson SP, Huertas P, Cortés-Ledesma F, Aguilera A. Muñoz-Galván S, et al. Nucleic Acids Res. 2013 Feb 1;41(3):1669-83. doi: 10.1093/nar/gks1274. Epub 2012 Dec 18. Nucleic Acids Res. 2013. PMID: 23254329 Free PMC article. - Small Molecule Inhibitors Confirm Ubiquitin-Dependent Removal of TOP2-DNA Covalent Complexes.
Swan RL, Poh LLK, Cowell IG, Austin CA. Swan RL, et al. Mol Pharmacol. 2020 Sep;98(3):222-233. doi: 10.1124/mol.119.118893. Epub 2020 Jun 25. Mol Pharmacol. 2020. PMID: 32587095 Free PMC article.
References
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. - PubMed
- Bergerat A, et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. - PubMed
- Connelly JC, Leach DR. Repair of DNA covalently linked to protein. Mol Cell. 2004;13:307–316. - PubMed
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. - PubMed
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases