Combined evidence annotation of transposable elements in genome sequences - PubMed (original) (raw)
Combined evidence annotation of transposable elements in genome sequences
Hadi Quesneville et al. PLoS Comput Biol. 2005 Jul.
Abstract
Transposable elements (TEs) are mobile, repetitive sequences that make up significant fractions of metazoan genomes. Despite their near ubiquity and importance in genome and chromosome biology, most efforts to annotate TEs in genome sequences rely on the results of a single computational program, RepeatMasker. In contrast, recent advances in gene annotation indicate that high-quality gene models can be produced from combining multiple independent sources of computational evidence. To elevate the quality of TE annotations to a level comparable to that of gene models, we have developed a combined evidence-model TE annotation pipeline, analogous to systems used for gene annotation, by integrating results from multiple homology-based and de novo TE identification methods. As proof of principle, we have annotated "TE models" in Drosophila melanogaster Release 4 genomic sequences using the combined computational evidence derived from RepeatMasker, BLASTER, TBLASTX, all-by-all BLASTN, RECON, TE-HMM and the previous Release 3.1 annotation. Our system is designed for use with the Apollo genome annotation tool, allowing automatic results to be curated manually to produce reliable annotations. The euchromatic TE fraction of D. melanogaster is now estimated at 5.3% (cf. 3.86% in Release 3.1), and we found a substantially higher number of TEs (n = 6,013) than previously identified (n = 1,572). Most of the new TEs derive from small fragments of a few hundred nucleotides long and highly abundant families not previously annotated (e.g., INE-1). We also estimated that 518 TE copies (8.6%) are inserted into at least one other TE, forming a nest of elements. The pipeline allows rapid and thorough annotation of even the most complex TE models, including highly deleted and/or nested elements such as those often found in heterochromatic sequences. Our pipeline can be easily adapted to other genome sequences, such as those of the D. melanogaster heterochromatin or other species in the genus Drosophila.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
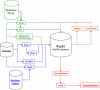
Figure 1. Schematic of Our TE Annotation Pipeline
The pipeline is composed of (i) known TE family detection methods such as BLRn, RM, and RMBLR; (ii) satellite detection software such as RM, TRF, and Mreps; (iii) anonymous TE detection methods such BLRa, TE-HMM, RECON, and BLRtx; and (iv) a MySQL database called REPET to manage the results and the annotations. GAME-XML files are then generated from the results stored in the database and loaded into the Apollo genome annotation tool, allowing automatic results to be manually curated to produce a reliable annotation. To facilitate manual curation, we automatically promoted RMBLR results to be the candidate annotation.
Figure 2. Screenshot of an Apollo View for a Peri-Centromeric Region with Extreme TE Density
Curated annotations on both forward strand (top) and reverse strand (bottom) are displayed in the light blue panels. Evidence tiers are shown in the black panels: TE-HMM (yellow), RECON (light purple), BLRa (violet), BLRtx (red), BLRn (teal), RM (blue), RMBLR (light green), and Release 3.1 FlyBase annotations (peach).
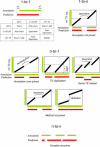
Figure 3. Categories of Possible Boundary Comparisons between Predictions and Reference Annotations
The different cases taken into account can be grouped according to one-to-one (1-to-1), one-to-many (1-to-n), many-to-one (_n_-to-1), or many-to-many (_n_-to-n) relationships.
Similar articles
- Discovering and detecting transposable elements in genome sequences.
Bergman CM, Quesneville H. Bergman CM, et al. Brief Bioinform. 2007 Nov;8(6):382-92. doi: 10.1093/bib/bbm048. Epub 2007 Oct 10. Brief Bioinform. 2007. PMID: 17932080 Review. - Improved detection and annotation of transposable elements in sequenced genomes using multiple reference sequence sets.
Buisine N, Quesneville H, Colot V. Buisine N, et al. Genomics. 2008 May;91(5):467-75. doi: 10.1016/j.ygeno.2008.01.005. Epub 2008 Mar 14. Genomics. 2008. PMID: 18343092 - A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes.
Kurtz S, Narechania A, Stein JC, Ware D. Kurtz S, et al. BMC Genomics. 2008 Oct 31;9:517. doi: 10.1186/1471-2164-9-517. BMC Genomics. 2008. PMID: 18976482 Free PMC article. - Colonization of heterochromatic genes by transposable elements in Drosophila.
Dimitri P, Junakovic N, Arcà B. Dimitri P, et al. Mol Biol Evol. 2003 Apr;20(4):503-12. doi: 10.1093/molbev/msg048. Epub 2003 Mar 5. Mol Biol Evol. 2003. PMID: 12654931 - Drosophila melanogaster: a case study of a model genomic sequence and its consequences.
Ashburner M, Bergman CM. Ashburner M, et al. Genome Res. 2005 Dec;15(12):1661-7. doi: 10.1101/gr.3726705. Genome Res. 2005. PMID: 16339363 Review.
Cited by
- Identification of transposable element families from pangenome polymorphisms.
Sierra P, Durbin R. Sierra P, et al. Mob DNA. 2024 Jun 26;15(1):13. doi: 10.1186/s13100-024-00323-y. Mob DNA. 2024. PMID: 38926873 Free PMC article. - On the Importance to Acknowledge Transposable Elements in Epigenomic Analyses.
Lerat E, Casacuberta J, Chaparro C, Vieira C. Lerat E, et al. Genes (Basel). 2019 Mar 31;10(4):258. doi: 10.3390/genes10040258. Genes (Basel). 2019. PMID: 30935103 Free PMC article. Review. - Functional insights from the GC-poor genomes of two aphid parasitoids, Aphidius ervi and Lysiphlebus fabarum.
Dennis AB, Ballesteros GI, Robin S, Schrader L, Bast J, Berghöfer J, Beukeboom LW, Belghazi M, Bretaudeau A, Buellesbach J, Cash E, Colinet D, Dumas Z, Errbii M, Falabella P, Gatti JL, Geuverink E, Gibson JD, Hertaeg C, Hartmann S, Jacquin-Joly E, Lammers M, Lavandero BI, Lindenbaum I, Massardier-Galata L, Meslin C, Montagné N, Pak N, Poirié M, Salvia R, Smith CR, Tagu D, Tares S, Vogel H, Schwander T, Simon JC, Figueroa CC, Vorburger C, Legeai F, Gadau J. Dennis AB, et al. BMC Genomics. 2020 May 29;21(1):376. doi: 10.1186/s12864-020-6764-0. BMC Genomics. 2020. PMID: 32471448 Free PMC article. - Deep investigation of Arabidopsis thaliana junk DNA reveals a continuum between repetitive elements and genomic dark matter.
Maumus F, Quesneville H. Maumus F, et al. PLoS One. 2014 Apr 7;9(4):e94101. doi: 10.1371/journal.pone.0094101. eCollection 2014. PLoS One. 2014. PMID: 24709859 Free PMC article. - Teaching transposon classification as a means to crowd source the curation of repeat annotation - a tardigrade perspective.
Peona V, Martelossi J, Almojil D, Bocharkina J, Brännström I, Brown M, Cang A, Carrasco-Valenzuela T, DeVries J, Doellman M, Elsner D, Espíndola-Hernández P, Montoya GF, Gaspar B, Zagorski D, Hałakuc P, Ivanovska B, Laumer C, Lehmann R, Boštjančić LL, Mashoodh R, Mazzoleni S, Mouton A, Nilsson MA, Pei Y, Potente G, Provataris P, Pardos-Blas JR, Raut R, Sbaffi T, Schwarz F, Stapley J, Stevens L, Sultana N, Symonova R, Tahami MS, Urzì A, Yang H, Yusuf A, Pecoraro C, Suh A. Peona V, et al. Mob DNA. 2024 May 6;15(1):10. doi: 10.1186/s13100-024-00319-8. Mob DNA. 2024. PMID: 38711146 Free PMC article.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
- Kidwell MG, Lisch DR. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution Int J Org Evolution. 2001;55:1–24. - PubMed
- Juretic N, Bureau TE, Bruskiewich RM. Transposable element annotation of the rice genome. Bioinformatics. 2004;20:155–160. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials