An in vitro approach to test the possible role of candidate factors in the transcriptional regulation of the RET proto-oncogene - PubMed (original) (raw)
An in vitro approach to test the possible role of candidate factors in the transcriptional regulation of the RET proto-oncogene
Tiziana Bachetti et al. Gene Expr. 2005.
Abstract
Neural crest cells arise from the epithelium of the dorsal neural tube and migrate to various districts giving origin, among others, to sympathetic, parasympathetic, and enteric ganglia. It has been shown that the transcription factors HOX11L1, HOX11L2, MASH1, PHOX2A, and PHOX2B are all necessary, to various extents, to the correct development of the autonomic nervous system. To investigate their possible role in the transcriptional regulation of the RET proto-oncogene, a gene playing a crucial role in correct intestinal innervation, we undertook a specific in vitro experimental strategy. Two neuroblastoma cell lines (SK-N-MC and SK-N-BE) were cotransfected with each transcription factor expressing plasmids and sequential deletion constructs of the 5' c-RET flanking region cloned upstream of the Luciferase reporter gene. Here we show that HOX11L1 enhances the activity of the c-RET promoter in SK-N-MC cell line by stimulating a region between -166 bp and -35 bp. Gel shift assays performed with oligonucleotides spanning this promoter sequence showed a change of the SP1 interaction with its binding sites, consequent to transfection with HOX11L1. While HOX11L2 showed no effect in both the cell lines, we have observed PHOX2A, PHOX2B, and MASH1 triggering a reproducible increase in the Luciferase activity in SK-N-BE cell line. A sequence responsible of the PHOX2A-dependent activation has been identified, while PHOX2B seems to act indirectly, as no physical binding has been demonstrated on c-RET promoter.
Figures
Figure 1
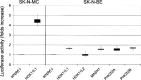
Cotransfections of the RET 5′ flanking region, cloned in a reporter gene vector with expression plasmids for either HOX11L1, HOX11L2, MASH1, PHOX2A, or PHOX2B, induce statistically significant increases of Luciferase activity expressed as folds of activation (p < 0.05) with respect to the empty expression vector (3.1TOPO). Each box represents median values and first and third quartile (error bars) of Luciferase activity. In the left panel, the result of the cotransfection performed in SK-N-MC of HOX11L1 with the RET promoter is reported, while the null effect of the other transcription factors tested in the same cell line on RET promoter is not shown. In the right panel, cotransfections of all the transcription factors with the RET promoter reporter construct are performed in SK-N-BE.
Figure 2
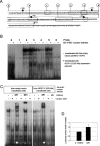
RET expression following transfection with HOX11L1. (A) SK-N-MC cells were transfected with pcDNA3.1TOPO-HOX11L1 (L1), with the empty vector (C−), and not transfected (PEI−). In any case, cells were treated with NaB to induce RET expression. RT-PCR was performed using a couple of primers designed on RET exon 18 and RET exon 20, respectively, and the amount of RET product compared to that obtained under the same condition for the GA3PDH gene. (B) Bands thus obtained were quantified to show the amount of RET expression in the presence and in the absence of HOX11L1. (C) Sequentially deleted reporter plasmids of the RET promoter, represented on the left, are reported along with corresponding HOX11L1 activations, plotted on the right as percentage of the activity of the full-length (−5078 bp) construct. Data represent the means ± SE (error bars) of at least three independent experiments performed in duplicate.
Figure 3
RET minimal promoter and interaction with nuclear extracts from SK-N-MC cells. (A) The sequence of the basal RET promoter has been divided into seven partially overlapping oligonucleotides numbered from 0 to 6. Arrows point to the starting nucleotide of the relative pGL3basic-RET PROMOTER deleted constructs used in cotransfection experiments with HOX11L1; the canonical SP1 recognition sites are written in italicized capital letters; +1 points to the transcription start site. (B) EMSA performed on a nondenaturing 5% polyacrylamide gel using seven γ-32P-labeled probes overlapping the entire sequence under analysis. (−) nuclear extract from SK-N-MC cells transfected with the empty vector; (+) nuclear extracts from pcDNA3.1TOPO-HOX11L1[V5-His] transfected SK-N-MC cells. (C) Supershift experiments with probe 3 and different antibodies: after incubation with SP1-specific antibody, a supershift is evident (white star) with nuclear extracts from both empty vector transfected (left panel) and HOX11L1 transfected (right panel) cells; in the left panel a typical retarded band for SP3 is shown (black star); each complex is specific because it disappeared after competition with a molar excess of cold probe. In nuclear extracts from pcDNA3.1TOPO-HOX11L1[V5-His] transfected cells, the incubation with the anti-V5 antibody did not produce any modification in the preformed complexes. (D) Cotransfection of the SP1 expression plasmid with pGL3basic-RET PROMOTER(5.1Kb) in SK-N-MC shows a mild increase in Luciferase activity, compared to the value obtained from the cotransfection with the empty vector (3.1 TOPO).
Figure 4
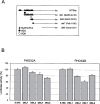
Cotransfection of PHOX2A and PHOX2B expression constructs with sequentially deleted fragments of the RET promoter. (A) Schematic distribution of TAAT boxes on the RET promoter. (B) Cotransfections of PHOX2A (left) and PHOX2B (right) expression constructs with reporter constructs containing regions of the RET promoter with a different number of TAAT boxes; cotransfections of the expression plasmids and the corresponding empty vector were performed in SK-N-BE and the diagrams are the result of three independent experiments performed in duplicate. For each RET promoter construct, the PHOX2 expression plasmid/empty vector ratios were calculated and values obtained with the deleted constructs expressed with respect to the full-length construct (100%), this latter corresponding to 1.6-fold activation induced by PHOX2A and PHOX2B (see Fig. 1).
Figure 5
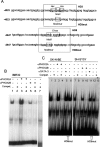
EMSA using oligonucleotides HD3 and HD8. (A) Sequences of the two regions of the 5′ RET flanking sequence considered in EMSA experiments. The putative homeodomain binding sites (TAAT) and the flanking regions included in the oligonucleotides are boxed and reported in both the wt (HD3, HD8) and the mutant (HD3mut, HD8mut) versions. (B) The 32P-labeled HD3 oligonucleotide was incubated with IMR32 nuclear extracts (lane 1); lane 2: IMR32 + unlabeled probe 200×; lane 3: IMR32 + αPHOX2A antibody; lane 4: IMR32 + αPHOX2A antibody unlabeled probe 200×; lane 5: IMR32 + αPHOX2B antibody; lane 6: IMR32 + αPHOX2B antibody unlabeled probe 200×; lane 7: IMR32 + unlabeled HD3mut probe. (C) The 32P-labeled HD8 oligonucleotide was incubated with 10 μg SK-N-BE nuclear extracts (lane 1); lane 2: SK-N-BE + unlabeled HD8 200×; lane 3: SK-N-BE + αPHOX2B; lane 4: SK-N-BE + αPHOX2A + unlabeled HD8 200×; lane 5: SK-N-BE + nonspecific antibody; lane 6: SK-N-BE nuclear extracts incubated with oligonucleotide HD8mut; lane 7: SH-SY5Y nuclear extracts; lane 8: SH-SY5Y + unlabeled HD8 200×; lane 9: SH-SY5Y + αPHOX2B; lane 10: SH-SY5Y + αPHOX2B + unlabeled HD8 200×; lane 11: SH-SY5Y + nonspecific antibody; lane 12: SH-SY5Y + incubated with oligonucleotide HD8mut, lane 13: SH-SY5Y + unlabeled HD8mut 200×.
Similar articles
- Sp1 and Sp3 transactivate the RET proto-oncogene promoter.
Andrew SD, Delhanty PJ, Mulligan LM, Robinson BG. Andrew SD, et al. Gene. 2000 Oct 3;256(1-2):283-91. doi: 10.1016/s0378-1119(00)00302-4. Gene. 2000. PMID: 11054558 - Transcriptional regulation of RET by Nkx2-1, Phox2b, Sox10, and Pax3.
Leon TY, Ngan ES, Poon HC, So MT, Lui VC, Tam PK, Garcia-Barcelo MM. Leon TY, et al. J Pediatr Surg. 2009 Oct;44(10):1904-12. doi: 10.1016/j.jpedsurg.2008.11.055. J Pediatr Surg. 2009. PMID: 19853745 - Sp proteins and Phox2b regulate the expression of the human Phox2a gene.
Flora A, Lucchetti H, Benfante R, Goridis C, Clementi F, Fornasari D. Flora A, et al. J Neurosci. 2001 Sep 15;21(18):7037-45. doi: 10.1523/JNEUROSCI.21-18-07037.2001. J Neurosci. 2001. PMID: 11549713 Free PMC article.
Cited by
- Role of RET and PHOX2B gene polymorphisms in risk of Hirschsprung's disease in Chinese population.
Miao X, Garcia-Barceló MM, So MT, Leon TY, Lau DK, Liu TT, Chan EK, Lan LC, Wong KK, Lui VC, Tam PK. Miao X, et al. Gut. 2007 May;56(5):736. doi: 10.1136/gut.2006.116145. Gut. 2007. PMID: 17440194 Free PMC article. No abstract available. - Chromosomal localization of PHOX2B during M-phase is disrupted in disease-associated mutants.
Sato Y, Hayashi S, Oe S, Koike T, Nakano Y, Seki-Omura R, Iwashita H, Hirahara Y, Kitada M. Sato Y, et al. Dev Growth Differ. 2025 Apr;67(3):136-148. doi: 10.1111/dgd.70001. Epub 2025 Feb 11. Dev Growth Differ. 2025. PMID: 39933489 Free PMC article. - Research Advances on Therapeutic Approaches to Congenital Central Hypoventilation Syndrome (CCHS).
Di Lascio S, Benfante R, Cardani S, Fornasari D. Di Lascio S, et al. Front Neurosci. 2021 Jan 12;14:615666. doi: 10.3389/fnins.2020.615666. eCollection 2020. Front Neurosci. 2021. PMID: 33510615 Free PMC article. Review. - Structural and functional differences in PHOX2B frameshift mutations underlie isolated or syndromic congenital central hypoventilation syndrome.
Di Lascio S, Benfante R, Di Zanni E, Cardani S, Adamo A, Fornasari D, Ceccherini I, Bachetti T. Di Lascio S, et al. Hum Mutat. 2018 Feb;39(2):219-236. doi: 10.1002/humu.23365. Epub 2017 Nov 21. Hum Mutat. 2018. PMID: 29098737 Free PMC article. - A novel bidirectional interaction between endothelin-3 and retinoic acid in rat enteric nervous system precursors.
Gisser JM, Cohen AR, Yin H, Gariepy CE. Gisser JM, et al. PLoS One. 2013 Sep 9;8(9):e74311. doi: 10.1371/journal.pone.0074311. eCollection 2013. PLoS One. 2013. PMID: 24040226 Free PMC article.
References
- Amiel J.; Laudier B.; Attie-Bitach T.; Trang H.; de Pontual L.; Gener B.; Trochet D.; Etchevers H.; Ray P.; Simonneau M.; Vekemans M.; Munnich A.; Gaultier C.; Lyonnet S. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 33:459–461; 2003. - PubMed
- Anderson D. J. Cellular and molecular biology of neural crest lineage determination. Trends Genet. 13:276–280; 1997. - PubMed
- Andreaw S. A.; Capes-Davis A.; Delhanty P. J. D.; Mash D. J.; Mulligan L. M.; Robinson B. G. Transcriptional repression of the RET proto-oncogene by a mitogen activated protein kinase-dependent signalling pathway. Gene 298:9–19; 2002. - PubMed
- Andreaw S. D.; Delhanty P. J. D.; Mulligan L. M.; Robinson B. G. Sp1 and Sp3 transactivate the RET proto-oncogene promoter. Gene 256:283–291; 2000. - PubMed
- Bouwman P.; Philipsen S. Regulation of the activity of SP1-related transcription factors. Mol. Cell. Endocrinol. 195:27–38; 2002. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources