Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A - PubMed (original) (raw)
Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A
Parker L Andersen et al. J Cell Biol. 2005.
Abstract
Ubc13, a ubiquitin-conjugating enzyme (Ubc), requires the presence of a Ubc variant (Uev) for polyubiquitination. Uevs, although resembling Ubc in sequence and structure, lack the active site cysteine residue and are catalytically inactive. The yeast Uev (Mms2) incites noncanonical Lys63-linked polyubiquitination by Ubc13, whereas the increased diversity of Uevs in higher eukaryotes suggests an unexpected complication in ubiquitination. In this study, we demonstrate that divergent activities of mammalian Ubc13 rely on its pairing with either of two Uevs, Uev1A or Mms2. Structurally, we demonstrate that Mms2 and Uev1A differentially modulate the length of Ubc13-mediated Lys63-linked polyubiquitin chains. Functionally, we describe that Ubc13-Mms2 is required for DNA damage repair but not nuclear factor kappaB (NF-kappaB) activation, whereas Ubc13-Uev1A is involved in NF-kappaB activation but not DNA repair. Our finding suggests a novel regulatory mechanism in which different Uevs direct Ubcs to diverse cellular processes through physical interaction and alternative polyubiquitination.
Figures
Figure 1.
Amino acid sequence comparison of yeast Mms2 and its human homologues. The amino acid sequences are obtained from the following sources: Mms2 (Broomfield et al., 1998); hMms2 (Xiao et al., 1998); Uev1A and Uev1B (Rothofsky and Lin, 1997). The corrected Uev1A sequence as reported previously (Deng et al., 2000) was used for alignment.
Figure 2.
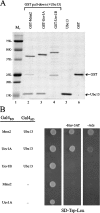
Ubc13–Uev complex formation. (A) In vitro GST pull-down assay. BL21(DE3)-RIL cells transformed with pGEX-hMMS2, pGEX-UEV1A, and pGEX-UEV1B were grown and target gene expression was induced by adding IPTG. Crude cell extracts were used to load GST affinity MicroSpin columns and 40 μg of purified hUbc13 was later added. The resulting elution was loaded onto an SDS-PAGE gel and stained with Coomassie blue. Components used in each sample are indicated on the top. Lane 5 contains 1 μg of purified hUbc13 and lane 6 contains 1 μg of purified GST, as indicated by arrows. (B) Yeast two-hybrid analysis of Ubc13–Uev interactions. Cotransformed plasmids are indicated on the left. The plates were incubated for 3 d before taking the photograph. Only one representative transformant from each combination is shown. Cells cotransformed with pUev1B-G4BD/pGAD424 and pG4BD-1/pGAD-Ubc13 were also negative (not depicted).
Figure 3.
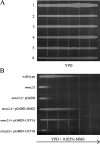
Heterologous function of human UEV genes in yeast. WXY903 (mms2_Δ::HIS3_) was transformed with various pUev-G4BD plasmids and the transformants were compared with HK580-10D (wild type) in a gradient plate assay for their ability to complement the yeast mms2 defect. Overnight cell cultures were printed on YPD (A) or YPD + 0.025% MMS (B) plates and incubated for 36 h before taking the photograph. The arrow points toward higher MMS concentration.
Figure 4.
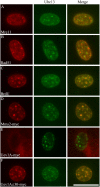
CPT-induced nuclear foci formation. ICC of 3T3 cells after CPT treatment (5 μM for 4 h) and in situ cell fractionation reveals Ubc13 nuclear foci (4E11 as primary antibody and Alexa488 as secondary antibody). These foci are compared, by merging images, with those of Mre11 (A), Rad51 (B), and BrdU (C) and in 3T3 cells transfected with Mms2-myc (D), Uev1A-myc (E), and Uev1AΔ30-myc (F) viewed by using specific primary antibodies and the Alexa546 secondary antibody, except in C, in which Alexa546 is directly conjugated with anti-BrdU. Bar, 5 μm.
Figure 5.
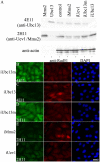
Spontaneous Rad51 nuclear foci formation after RNAi treatment. (A) Western blot analysis of purified recombinant hMms2 (lane 1), hUbc13 (lane 2), and 3T3 total cell extracts (lanes 3–7) using 4E11 (anti-Ubc13) or 2H11 (anti-hMms2) as primary antibodies. Lanes 4–7 represent cultures transfected with various RNAi constructs as indicated. Note that 2H11 (and other anti-hMms2 mAbs) detected only one band from mammalian cell extracts, which corresponds to the migration of Mms2, but was found to contain both Mms2 and Uev1A by mass spectrometry (not depicted). (B–F) 4 d after transfection of 3T3 cells with the indicated RNAi constructs against specific target genes, cells were stained with either 4E11 (B and C) or 2H11 (D–F) plus Alexa488 to reveal Ubc13 and Uev immunoreactivity, respectively. The same cells were also stained with anti-Rad51 plus Alexa546 to reveal Rad51 nuclear foci and with DAPI to reveal nuclei of all cells. Bar, 10 μm.
Figure 6.
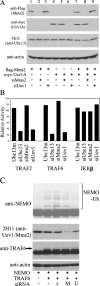
Ablation of Uev1A suppresses TRAF-induced NF-κB activity and NEMO polyubiquitination in 293T cells. (A) Western blot analysis confirms RNAi specificity and efficacy of target suppression. (B) Modulation of IKKβ-, TRAF2-, and TRAF6-induced NF-κB activity by ablation of Ubc13, Uev1, or Mms2. The relative NF-κB activity was calculated against transfected cells without siRNA treatment. (C) Effects of Mms2 or Uev1 ablation by siRNA on NEMO polyubiquitination. s, scrambled nonspecific siRNA; M, siMms2; U, siUev1. The arrow points to TRAF6.
Figure 7.
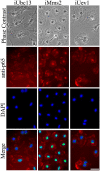
Requirement of Ubc13 and Uev1A for LPS-induced p65 translocation. Phase contrast, p65-ICC, and DAPI staining were performed 4 d after transfection of mouse microglia with RNAi constructs as indicated, followed by a 1.5-h exposure to 1 μg/ml LPS and fixation. Merged images indicate colocalization of p65 immunostaining with nuclei. Identical color adjustment was made to all merged images to enhance differential colocalization. Bar, 10 μm.
Figure 8.
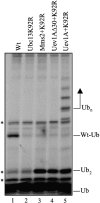
Ub chain building. Mms2 and Uev1A were assayed for Ub chain building activity using an in vitro ubiquitination reaction. The components of the reaction are noted in Materials and methods. The concentration of wild-type hUbc13 (Wt), hUbc13K92R (K92R), hMms2, Uev1A, and Uev1AΔ30 is 250 nM. The positions of free Ub (Ub), di-Ub (Ub2), Ubc13-Ub (Wt-Ub), and multi-Ub chains (Ubn) are indicated. Major contaminant bands from the 35S-Ub preparation, including the background band comigrating with Ub2, are indicated by an asterisk.
Figure 9.
A working model of Ubc13–Uev functions in human cells. This model is based on data and discussion presented in this paper as well as some previous papers. Please note that a mammalian Rad5 homologue has not been identified.
Similar articles
- Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation.
Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Eddins MJ, et al. Nat Struct Mol Biol. 2006 Oct;13(10):915-20. doi: 10.1038/nsmb1148. Epub 2006 Sep 17. Nat Struct Mol Biol. 2006. PMID: 16980971 - Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response.
Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W. Wen R, et al. Plant Cell. 2008 Jan;20(1):213-27. doi: 10.1105/tpc.107.051862. Epub 2008 Jan 4. Plant Cell. 2008. PMID: 18178771 Free PMC article. - A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex.
Pastushok L, Moraes TF, Ellison MJ, Xiao W. Pastushok L, et al. J Biol Chem. 2005 May 6;280(18):17891-900. doi: 10.1074/jbc.M410469200. Epub 2005 Mar 4. J Biol Chem. 2005. PMID: 15749714 - Ubc13: the Lys63 ubiquitin chain building machine.
Hodge CD, Spyracopoulos L, Glover JN. Hodge CD, et al. Oncotarget. 2016 Sep 27;7(39):64471-64504. doi: 10.18632/oncotarget.10948. Oncotarget. 2016. PMID: 27486774 Free PMC article. Review. - Functions and mechanisms of the Ubc13-UEV complex and lysine 63-linked polyubiquitination in plants.
Yang K, Xiao W. Yang K, et al. J Exp Bot. 2022 Sep 12;73(16):5372-5387. doi: 10.1093/jxb/erac239. J Exp Bot. 2022. PMID: 35640002 Review.
Cited by
- Rice UBC13, a candidate housekeeping gene, is required for K63-linked polyubiquitination and tolerance to DNA damage.
Zang Y, Wang Q, Xue C, Li M, Wen R, Xiao W. Zang Y, et al. Rice (N Y). 2012 Dec;5(1):24. doi: 10.1186/1939-8433-5-24. Epub 2012 Sep 8. Rice (N Y). 2012. PMID: 27234244 Free PMC article. - Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling.
Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, Nakaso K, Culmsee C, Berninger B, Krappmann D, Tatzelt J, Winklhofer KF. Henn IH, et al. J Neurosci. 2007 Feb 21;27(8):1868-78. doi: 10.1523/JNEUROSCI.5537-06.2007. J Neurosci. 2007. PMID: 17314283 Free PMC article. - Tizoxanide Antiviral Activity on Dengue Virus Replication.
Yamamoto KA, Blackburn K, Goshe MB, Brown DT, Migoswski E, Campanhon IB, Moreira MF, Ferreira DF, Soares MR. Yamamoto KA, et al. Viruses. 2023 Mar 7;15(3):696. doi: 10.3390/v15030696. Viruses. 2023. PMID: 36992406 Free PMC article. - UBC13 is an RNF213-associated E2 ubiquitin-conjugating enzyme, and Lysine 63-linked ubiquitination by the RNF213-UBC13 axis is responsible for angiogenic activity.
Habu T, Harada KH. Habu T, et al. FASEB Bioadv. 2021 Jan 20;3(4):243-258. doi: 10.1096/fba.2019-00092. eCollection 2021 Apr. FASEB Bioadv. 2021. PMID: 33842849 Free PMC article. - Advanced Cataloging of Lysine-63 Polyubiquitin Networks by Genomic, Interactome, and Sensor-Based Proteomic Analyses.
Romero-Barrios N, Monachello D, Dolde U, Wong A, San Clemente H, Cayrel A, Johnson A, Lurin C, Vert G. Romero-Barrios N, et al. Plant Cell. 2020 Jan;32(1):123-138. doi: 10.1105/tpc.19.00568. Epub 2019 Nov 11. Plant Cell. 2020. PMID: 31712406 Free PMC article.
References
- Andegeko, Y., L. Moyal, L. Mittelman, I. Tsarfaty, Y. Shiloh, and G. Rotman. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224–38230. - PubMed
- Bonaiuto, C., P.P. McDonald, F. Rossi, and M.A. Cassatella. 1997. Activation of nuclear factor-κB by β-amyloid peptides and interferon-γ in murine microglia. J. Neuroimmunol. 77:51–56. - PubMed
- Bothos, J., M.K. Summers, M. Venere, D.M. Scolnick, and T.D. Halazonetis. 2003. The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene. 22:7101–7107. - PubMed
- Brinkmann, U., M. Gallo, M.H. Polymeropoulos, and I. Pastan. 1996. The human CAS (cellular apoptosis susceptibility) gene mapping on chromosome 20q13 is amplified in BT474 breast cancer cells and part of aberrant chromosomes in breast and colon cancer cell lines. Genome Res. 6:187–194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous