Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis - PubMed (original) (raw)
Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis
Dominique Pontier et al. Genes Dev. 2005.
Abstract
Recent genetic and biochemical studies have revealed the existence in plants of a fourth RNA polymerase, RNAPIV, which mediates siRNA accumulation and DNA methylation-dependent silencing of endogenous repeated sequences. Here, we show that Arabidopsis expresses, in fact, two evolutionarily related forms of RNAPIV, hereafter referred to as RNAPIVa and RNAPIVb. These two forms contain the same second-largest subunit (NRPD2), but differ at least by their largest subunit, termed NRPD1a and NRPD1b. Unlike NRPD1a, NRPD1b possesses a reiterated CTD, a feature that also characterizes the largest subunit of RNAPII. Our data indicate that RNAPIVb is the most abundant form of RNAPIV in Arabidopsis. Selective disruption of either form of RNAPIV indicates that RNAPIVa-dependent siRNA accumulation is not sufficient per se to drive robust silencing at endogenous loci and that high levels of DNA methylation and silencing depend on siRNA that are accumulated through a pathway involving the concerted action of both RNAPIV forms. Taken together, our results imply the existence of a novel two-step mechanism in siRNA synthesis at highly methylated loci, with RNAPIVb being an essential component of a self-reinforcing loop coupling de novo DNA methylation to siRNA production.
Figures
Figure 1.
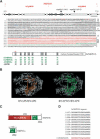
Arabidopsis contains a second class IV largest subunit gene. (A) Diagram of AtNRPD1b gene and corresponding protein product. Predicted and reannotated exons are indicated with open and gray boxes, respectively. Vertical arrowheads indicate T-DNA insertions. Evolutionarily conserved regions A to H are represented as gray boxes. The cysteine and histidine residues of the zinc-binding domain in the conserved region A are indicated in red (cc). The catalytic aspartate residues present in the conserved region D are indicated in blue. The hydrophilic S/G/A/D/E/K-rich region and the DCL-like domains that compose the CTD are red and green, respectively. Reiterated motifs are underlined. Numbers refer to amino acid identities between AtNRPD1b conserved regions and the corresponding domains in homologs corresponding to SoNRPD1b, OsNRPD1b, AtNRPD1a, and AtRPB1. (B) Conservation of an RNA polymerase core structure in RNAPIVb. The active-site magnesium is indicated by a pink sphere. (C) Schematic structure of AtNRPD1b and AtNRPD1a CTDs. The S/G/A/D/E/K-rich region is shown in red, and the DCL-like domains are shown in green. Repeated motifs are indicated as vertical lines. (D) Amino acid alignment of the repeated motifs with the deduced consensus sequence.
Figure 2.
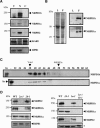
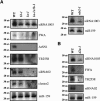
Arabidopsis contains two RNAPIV forms. (A) Western blot analysis of Arabidopsis proteins from total extracts (T), percoll-purified nuclei (N), and chloroplasts (C) with NRPD1b, NRPD1a, NRPD2, KARI, and RPB1 antibodies. KARI was used as a plastid control protein, whereas RPB1 was used as a nuclear control. (B) Western blot analysis and Coomassie blue staining of leaf (L) and flower (F) extracts (∼20 μg) with NRPD1a and NRPD1b antibodies. (C) Gel filtration chromatographic analysis of the NRPD1-containing complexes. The eluted fractions were analyzed by Western blots using the NRPD1b and NRPD1a antibodies. Numbered lanes correspond to size-fractionated protein fractions ranging from 250 kDa (lane 58) to 2 MDa (lane 38). The peak position of the ferritin protein marker (440 kDa) is indicated. (In) Input material. (D) Mutual stability of the class IV largest and second-largest subunits in various null mutants. The steady state of NRPD1b and NRPD1a was analyzed in various nrpd2 mutants (left panel). RPB1 protein was used as a loading control. The steady state of NRPD2 was analyzed in various nrpd1 mutants (right panel). A nonspecific cross-reacting band indicated by an asterisk was used as a loading control.
Figure 3.
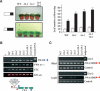
Phenotypes of nrpd1 mutant lines. (A) Phenotype of nrpd1 plants, grown under long days (top) and short days (bottom, left). The picture represents one out of nine plants per line. (Right) Comparison of the leaf number at flowering of wild-type (WT) and nrpd1 single and double mutants. Similar results were obtained for 1a-2 and 1b-2 mutants. (B) RT–PCR analysis of flowering-related gene expression in homozygous single and double mutants. The FWA panel corresponds to the amplification of the coding region spanning exons 3–5 of the gene. The _FWA_tr corresponds to the tandem repeats located upstream of FWA. The cartoon shows the location of _FWA_-derived primer pairs used in panels B and C. (C) Analysis of DNA methylation of the FWA tandem repeats. DNA was digested with the methylation-sensitive enzymes HhaI (top) and AvaII (bottom).
Figure 4.
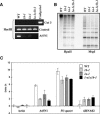
RNAPIVa and RNAPIVb act nonredundantly in the same pathway controlling methylation at AtSN1 and 5s rDNA. (A) Analysis of DNA methylation of AtSN1. HaeIII-digested DNA was used as a template for PCR reactions using AtSN1 and control primers. The cut3 primers would not amplify DNA if the digestion were complete. Undigested corresponds to undigested DNA. (B) Blot analysis of 5S rDNA digested with methylation-sensitive restriction enzymes HpaII (left) and MspI (right) in nrpd1 mutants and hybridized to a 5S probe. (C) DNA methylation at various repeated loci was assessed in different mutant backgrounds by quantitative McrBC–PCR. Delta Ct corresponds to the difference in Ct between digested and undigested samples. Heavily methylated sequences give large Delta Ct, whereas unmethylated sequences give a Delta Ct value of 0.
Figure 5.
Differential role of the two RNAPIVs on siRNA accumulation. (A) Small RNA blot assays for miR-159 and various endogenous siRNAs in various nrpd1 mutants. Blots in A were stripped and reprobed multiple times as indicated on the right. (Right panel) Small RNA blot assays for miR-159 and siRNA 1003 in the rdr2-1 mutant is shown. (B) Small RNA blot assays for miR-159 and various endogenous siRNAs in nrpd2a-1 mutant.
Figure 6.
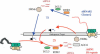
Hypothetical model for siRNA-dependent DNA methylation at endogenous repeated loci in plants. The blue arrows correspond to the RNAPIVa-dependent pathway that concerns siRNA02 and cluster 2. On targets such as AtSN1 and 5S repeats it would be followed by a self-reinforcing loop implicating RNAPIVb (red arrows).
Similar articles
- RNA polymerase IV directs silencing of endogenous DNA.
Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. Herr AJ, et al. Science. 2005 Apr 1;308(5718):118-20. doi: 10.1126/science.1106910. Epub 2005 Feb 3. Science. 2005. PMID: 15692015 - NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis.
Eamens A, Vaistij FE, Jones L. Eamens A, et al. Plant J. 2008 Aug;55(4):596-606. doi: 10.1111/j.1365-313X.2008.03525.x. Epub 2008 Apr 22. Plant J. 2008. PMID: 18433438 - RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants.
Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. Huettel B, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):358-74. doi: 10.1016/j.bbaexp.2007.03.001. Epub 2007 Mar 12. Biochim Biophys Acta. 2007. PMID: 17449119 Review. - Finding the right template: RNA Pol IV, a plant-specific RNA polymerase.
Vaughn MW, Martienssen RA. Vaughn MW, et al. Mol Cell. 2005 Mar 18;17(6):754-6. doi: 10.1016/j.molcel.2005.03.003. Mol Cell. 2005. PMID: 15780931 Review.
Cited by
- Abundant expression of maternal siRNAs is a conserved feature of seed development.
Grover JW, Burgess D, Kendall T, Baten A, Pokhrel S, King GJ, Meyers BC, Freeling M, Mosher RA. Grover JW, et al. Proc Natl Acad Sci U S A. 2020 Jun 30;117(26):15305-15315. doi: 10.1073/pnas.2001332117. Epub 2020 Jun 15. Proc Natl Acad Sci U S A. 2020. PMID: 32541052 Free PMC article. - Parallel action of AtDRB2 and RdDM in the control of transposable element expression.
Clavel M, Pélissier T, Descombin J, Jean V, Picart C, Charbonel C, Saez-Vásquez J, Bousquet-Antonelli C, Deragon JM. Clavel M, et al. BMC Plant Biol. 2015 Mar 3;15:70. doi: 10.1186/s12870-015-0455-z. BMC Plant Biol. 2015. PMID: 25849103 Free PMC article. - The non-coding RNA SVALKA locus produces a cis-natural antisense transcript that negatively regulates the expression of CBF1 and biomass production at normal temperatures.
Zacharaki V, Meena SK, Kindgren P. Zacharaki V, et al. Plant Commun. 2023 Jul 10;4(4):100551. doi: 10.1016/j.xplc.2023.100551. Epub 2023 Jan 21. Plant Commun. 2023. PMID: 36681861 Free PMC article. - RNA-directed DNA methylation: mechanisms and functions.
Mahfouz MM. Mahfouz MM. Plant Signal Behav. 2010 Jul;5(7):806-16. doi: 10.4161/psb.5.7.11695. Epub 2010 Jul 1. Plant Signal Behav. 2010. PMID: 20421728 Free PMC article. Review. - Ancient origin, functional conservation and fast evolution of DNA-dependent RNA polymerase III.
Proshkina GM, Shematorova EK, Proshkin SA, Zaros C, Thuriaux P, Shpakovski GV. Proshkina GM, et al. Nucleic Acids Res. 2006 Jul 28;34(13):3615-24. doi: 10.1093/nar/gkl421. Print 2006. Nucleic Acids Res. 2006. PMID: 16877568 Free PMC article.
References
- Allison L.A., Moyle, M., Shales, M., and Ingles, C.J. 1985. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 42: 599-610. - PubMed
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, D.K., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. - PubMed
- Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. - PubMed
- Bentley D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11: 347-351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases