Fungal community analysis by large-scale sequencing of environmental samples - PubMed (original) (raw)
Fungal community analysis by large-scale sequencing of environmental samples
Heath E O'Brien et al. Appl Environ Microbiol. 2005 Sep.
Abstract
Fungi are an important and diverse component of soil communities, but these communities have proven difficult to study in conventional biotic surveys. We evaluated soil fungal diversity at two sites in a temperate forest using direct isolation of small-subunit and internal transcribed spacer (ITS) rRNA genes by PCR and high-throughput sequencing of cloned fragments. We identified 412 sequence types from 863 fungal ITS sequences, as well as 112 ITS sequences from other eukaryotic microorganisms. Equal proportions of Basidiomycota and Ascomycota sequences were present in both the ITS and small-subunit libraries, while members of other fungal phyla were recovered at much lower frequencies. Many sequences closely matched sequences from mycorrhizal, plant-pathogenic, and saprophytic fungi. Compositional differences were observed among samples from different soil depths, with mycorrhizal species predominating deeper in the soil profile and saprophytic species predominating in the litter layer. Richness was consistently lowest in the deepest soil horizon samples. Comparable levels of fungal richness have been observed following traditional specimen-based collecting and culturing surveys, but only after much more extensive sampling. The high rate at which new sequence types were recovered even after sampling 863 fungal ITS sequences and the dominance of fungi in our libraries relative to other eukaryotes suggest that the abundance and diversity of fungi in forest soils may be much higher than previously hypothesized. All sequences were deposited in GenBank, with accession numbers AY 969316 to AY 970290 for the ITS sequences and AY 969135 to AY 969315 for the SSU sequences.
Figures
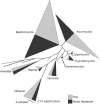
FIG. 1.
Proportions of SSU sequences belonging to different fungal phyla and other eukaryotic kingdoms. The width of triangles corresponds to the number of sequences in each, and the height corresponds to the maximum amount of sequence divergence within clades (see scale bars).
FIG. 2.
Proportional distribution of different taxonomic groups in ITS clone libraries. The x axis indicates the proportion of sequences assigned to each taxonomic group, and bar width corresponds to the proportion of all sequences belonging to each library. A. Proportion of all ITS sequences belonging to each fungal kingdom. B. Proportion of Ascomycota ITS sequences belonging to each subclass. C. Proportion of Basidiomycota ITS sequences belonging to each order.
FIG. 3.
Species-effort curves of richness for fungal OTUs. A. Mixed hardwood plot. B. Pine plot. C. Pooled data from both plots.
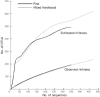
FIG. 4.
Species-effort curves for observed and estimated fungal OTU richness in each plot.
Similar articles
- Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil.
Anderson IC, Campbell CD, Prosser JI. Anderson IC, et al. Environ Microbiol. 2003 Jan;5(1):36-47. doi: 10.1046/j.1462-2920.2003.00383.x. Environ Microbiol. 2003. PMID: 12542711 - 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity.
Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F. Buée M, et al. New Phytol. 2009 Oct;184(2):449-456. doi: 10.1111/j.1469-8137.2009.03003.x. Epub 2009 Aug 22. New Phytol. 2009. PMID: 19703112 - Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR.
Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. Arnold AE, et al. Mycologia. 2007 Mar-Apr;99(2):185-206. doi: 10.3852/mycologia.99.2.185. Mycologia. 2007. PMID: 17682771 - Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis.
Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K. Smit E, et al. Appl Environ Microbiol. 1999 Jun;65(6):2614-21. doi: 10.1128/AEM.65.6.2614-2621.1999. Appl Environ Microbiol. 1999. PMID: 10347051 Free PMC article. - Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi.
Wu B, Hussain M, Zhang W, Stadler M, Liu X, Xiang M. Wu B, et al. Mycology. 2019 May 7;10(3):127-140. doi: 10.1080/21501203.2019.1614106. eCollection 2019. Mycology. 2019. PMID: 31448147 Free PMC article. Review.
Cited by
- Accurate Estimation of Fungal Diversity and Abundance through Improved Lineage-Specific Primers Optimized for Illumina Amplicon Sequencing.
Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, Caporaso JG, Pennanen T. Taylor DL, et al. Appl Environ Microbiol. 2016 Nov 21;82(24):7217-7226. doi: 10.1128/AEM.02576-16. Print 2016 Dec 15. Appl Environ Microbiol. 2016. PMID: 27736792 Free PMC article. - Filling gaps in biodiversity knowledge for macrofungi: contributions and assessment of an herbarium collection DNA barcode sequencing project.
Osmundson TW, Robert VA, Schoch CL, Baker LJ, Smith A, Robich G, Mizzan L, Garbelotto MM. Osmundson TW, et al. PLoS One. 2013 Apr 30;8(4):e62419. doi: 10.1371/journal.pone.0062419. Print 2013. PLoS One. 2013. PMID: 23638077 Free PMC article. - Eosinophils in fungus-associated allergic pulmonary disease.
Ghosh S, Hoselton SA, Dorsam GP, Schuh JM. Ghosh S, et al. Front Pharmacol. 2013 Feb 1;4:8. doi: 10.3389/fphar.2013.00008. eCollection 2013. Front Pharmacol. 2013. PMID: 23378838 Free PMC article. - Short-term temporal variability in airborne bacterial and fungal populations.
Fierer N, Liu Z, Rodríguez-Hernández M, Knight R, Henn M, Hernandez MT. Fierer N, et al. Appl Environ Microbiol. 2008 Jan;74(1):200-7. doi: 10.1128/AEM.01467-07. Epub 2007 Nov 2. Appl Environ Microbiol. 2008. PMID: 17981945 Free PMC article. - A High-Resolution Time Series Reveals Distinct Seasonal Patterns of Planktonic Fungi at a Temperate Coastal Ocean Site (Beaufort, North Carolina, USA).
Duan Y, Xie N, Song Z, Ward CS, Yung CM, Hunt DE, Johnson ZI, Wang G. Duan Y, et al. Appl Environ Microbiol. 2018 Oct 17;84(21):e00967-18. doi: 10.1128/AEM.00967-18. Print 2018 Nov 1. Appl Environ Microbiol. 2018. PMID: 30143506 Free PMC article.
References
- Anderson, I. C., C. D. Campbell, and J. I. Prosser. 2003. Diversity of fungi in organic soils under a moorland-Scots pine (Pinus sylvestris L.) gradient. Environ. Microbiol. 5:1121-1132. - PubMed
- Arnold, A. E., Z. Maynard, G. S. Gilbert, P. D. Coley, and T. A. Kursar. 2000. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 3:267-274.
- Bridge, P. D., and B. M. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154.
- Bruns, T. D., T. J. White, and J. W. Taylor. 1991. Fungal molecular systematics. Annu. Rev. Ecol. Syst. 22:525-564.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases