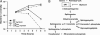Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen - PubMed (original) (raw)
Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen
Luisa W Cheng et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Three genome-wide RNA interference screens were performed in Drosophila S2 cells to dissect the contribution of host processes to Listeria monocytogenes entry, vacuolar escape, and intracellular growth. Among the 116 genes identified, several host pathways previously unrecognized as playing a role in listerial pathogenesis were identified: knockdowns affecting vacuolar trafficking to and from the multivesicular body bypassed the requirement for the essential pore-forming toxin listeriolysin O in mediating escape from phagocytic vacuoles and knockdowns affecting either subunit of serine palmitoyltransferase, a key enzyme in ceramide and sphingolipid biosynthesis, enhanced the toxicity of listeriolysin O expressed in the host cell cytosol, leading to lack of appropriate toxin activity compartmentalization and host cell death. Genome-wide RNA interference screens using Drosophila S2 cells proved to be a powerful approach to dissect host-pathogen interactions.
Figures
Fig. 1.
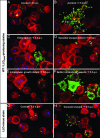
Immunofluorescence micrographs of S2 cells infected with L. monocytogenes strains. Bacteria were labeled by using a rabbit polyclonal anti-Listeria antibody followed by secondary labeling with an Alexa 488-coupled anti-rabbit antibody (shown in green). S2 cells and actin were stained in red with tetramethylrhodamine B isothiocyanate-phalloidin, and cell nuclei were stained in blue with DAPI. Phenotypes for S2 cells infected with wild-type bacteria and the LLOS44A-producing strain were very similar; herein, only micrographs of cells infected with wild-type L. monocytogenes are presented (A-F). (A and B) Control cells (no RNAi treatment) at 2 and 7.5 h postinfection (h.p.i.), respectively. (C-H) Micrographs depict RNAi phenotypes at 7.5 h.p.i. (C) Entry defect when chc (clathrin heavy chain) was silenced. (D) Defect in vacuolar escape when vha13 (vATPAse subunit) was silenced; more bacteria are seen as “clumps.” (E) Defect in intracellular growth when peanut (Septin 7 homologue) was silenced. (F) Better intracellular growth when CG5451 was silenced. (G) Control cells infected with LLO-minus bacteria; LLO-minus is defective in vacuolar escape. (H) Vacuolar escape phenotype of the LLO-minus strain when dor (Vps18 homologue) was silenced; here a knockdown bypassed the requirement for LLO in vacuolar escape.
Fig. 2.
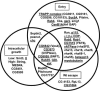
RNAi affecting L. monocytogenes entry, escape, and intracellular growth. Diagram illustrating the interconnecting phenotypes of select genes identified in the wild-type L. monocytogenes RNAi screen. Genes are categorized based on phenotypes for entry, vacuolar escape, and intracellular growth. Underlined are genes involved in protein or vesicular trafficking.
Fig. 3.
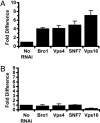
RNAi knockdowns that bypass vacuolar escape defect of LLO-minus L. monocytogenes. (A) Vacuolar escape of LLO-minus bacteria. In no-RNAi-treated S2 cells, LLO-minus bacteria escaped vacuoles at an average of 2 ± 0.4%. Vacuolar escape of knockdown mutants was represented as fold difference of the no-RNAi-treated control (set at the arbitrary unit of 1). Vacuolar escape was calculated as described in Materials and Methods. (B) Vacuolar escape of LLO&PLC-minus bacteria. LLO&PLC-minus mutant escaped at an average of 0.7 ± 0.1%.
Fig. 4.
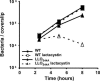
Intracellular growth of wild-type and LLOS44A-producing strains in Drosophila S2 cells upon inhibition of the proteasome. Inhibition of the proteasome with 2.5 μM lactacystin (dashed line) increased toxicity caused by the LLOS44A-producing strain (triangle). Growth of wild-type bacteria (square) was not affected by the addition of lactacystin. Gentamicin was added to the medium 1 h postinfection to kill extracellular bacteria and bacteria in host cells whose plasma membrane was compromised. Decrease in intracellular growth of bacteria is correlated with the degree of LLO toxicity.
Fig. 5.
Effect of SPT inhibition on bacterial intracellular growth in murine bone marrow-derived macrophages. (A) Growth of wild-type L. monocytogenes (square) and LLOS44A-producing strains (circle) in murine bone marrow-derived macrophages, upon inhibition of SPT with 10 μM myriocin (dashed lines). Inhibition of SPT increased toxicity of the LLOS44A-producing strain leading to the increased influx of gentamicin into the cytosol and subsequent killing of intracellular bacteria. (B) Mammalian sphingolipid and catabolism pathways. Myriocin is an inhibitor of SPT.
Similar articles
- Listeria monocytogenes exploits cystic fibrosis transmembrane conductance regulator (CFTR) to escape the phagosome.
Radtke AL, Anderson KL, Davis MJ, DiMagno MJ, Swanson JA, O'Riordan MX. Radtke AL, et al. Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1633-8. doi: 10.1073/pnas.1013262108. Epub 2011 Jan 10. Proc Natl Acad Sci U S A. 2011. PMID: 21220348 Free PMC article. - Drosophila S2 cells: an alternative infection model for Listeria monocytogenes.
Cheng LW, Portnoy DA. Cheng LW, et al. Cell Microbiol. 2003 Dec;5(12):875-85. doi: 10.1046/j.1462-5822.2003.00327.x. Cell Microbiol. 2003. PMID: 14641173 - Relative Roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes Uptake by Host Cells.
Phelps CC, Vadia S, Arnett E, Tan Y, Zhang X, Pathak-Sharma S, Gavrilin MA, Seveau S. Phelps CC, et al. Infect Immun. 2018 Sep 21;86(10):e00555-18. doi: 10.1128/IAI.00555-18. Print 2018 Oct. Infect Immun. 2018. PMID: 30061379 Free PMC article. - The molecular mechanisms of listeriolysin O-induced lipid membrane damage.
Petrišič N, Kozorog M, Aden S, Podobnik M, Anderluh G. Petrišič N, et al. Biochim Biophys Acta Biomembr. 2021 Jul 1;1863(7):183604. doi: 10.1016/j.bbamem.2021.183604. Epub 2021 Mar 17. Biochim Biophys Acta Biomembr. 2021. PMID: 33722646 Review. - Listeriolysin O: a phagosome-specific lysin.
Schnupf P, Portnoy DA. Schnupf P, et al. Microbes Infect. 2007 Aug;9(10):1176-87. doi: 10.1016/j.micinf.2007.05.005. Epub 2007 May 7. Microbes Infect. 2007. PMID: 17720603 Review.
Cited by
- Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference.
Li X, Dong X, Zou C, Zhang H. Li X, et al. Sci Rep. 2015 Mar 3;5:8700. doi: 10.1038/srep08700. Sci Rep. 2015. PMID: 25731667 Free PMC article. - Host-Targeted Therapeutics against Multidrug Resistant Intracellular Staphylococcus aureus.
Bravo-Santano N, Behrends V, Letek M. Bravo-Santano N, et al. Antibiotics (Basel). 2019 Nov 28;8(4):241. doi: 10.3390/antibiotics8040241. Antibiotics (Basel). 2019. PMID: 31795127 Free PMC article. Review. - Cytosolic detection of phagosomal bacteria-Mechanisms underlying PAMP exodus from the phagosome into the cytosol.
Ragland SA, Kagan JC. Ragland SA, et al. Mol Microbiol. 2021 Dec;116(6):1420-1432. doi: 10.1111/mmi.14841. Epub 2021 Nov 22. Mol Microbiol. 2021. PMID: 34738270 Free PMC article. Review. - Identification of inhibitors of inositol 5-phosphatases through multiple screening strategies.
Pirruccello M, Nandez R, Idevall-Hagren O, Alcazar-Roman A, Abriola L, Berwick SA, Lucast L, Morel D, De Camilli P. Pirruccello M, et al. ACS Chem Biol. 2014 Jun 20;9(6):1359-68. doi: 10.1021/cb500161z. Epub 2014 May 1. ACS Chem Biol. 2014. PMID: 24742366 Free PMC article. - Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions?
Cherry S. Cherry S. Curr Opin Microbiol. 2008 Jun;11(3):262-70. doi: 10.1016/j.mib.2008.05.007. Epub 2008 Jun 6. Curr Opin Microbiol. 2008. PMID: 18539520 Free PMC article. Review.
References
- Alonso, A. & Garcia-del Portillo, F. (2004) Int. Microbiol. 7, 181-191. - PubMed
- Decatur, A. L. & Portnoy, D. A. (2000) Science 290, 992-995. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI29619/AI/NIAID NIH HHS/United States
- AI27655/AI/NIAID NIH HHS/United States
- F32 AI051896/AI/NIAID NIH HHS/United States
- R01 AI027655/AI/NIAID NIH HHS/United States
- R37 AI029619/AI/NIAID NIH HHS/United States
- AI51896/AI/NIAID NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases