Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility - PubMed (original) (raw)
Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility
Laurence Nachin et al. J Bacteriol. 2005 Sep.
Abstract
The universal stress protein (UspA) superfamily encompasses a conserved group of proteins that are found in bacteria, archaea, and eukaryotes. Escherichia coli harbors six usp genes--uspA, -C, -D, -E, -F, and -G--the expression of which is triggered by a large variety of environmental insults. The uspA gene is important for survival during cellular growth arrest, but the exact physiological role of the Usp proteins is not known. In this work we have performed phenotypic characterization of mutants with deletions of the six different usp genes. We report on hitherto unknown functions of these genes linked to motility, adhesion, and oxidative stress resistance, and we show that usp functions are both overlapping and distinct. Both UspA and UspD are required in the defense against superoxide-generating agents, and UspD appears also important in controlling intracellular levels of iron. In contrast, UspC is not involved in stress resistance or iron metabolism but is essential, like UspE, for cellular motility. Electron microscopy demonstrates that uspC and uspE mutants are devoid of flagella. In addition, the function of the uspC and uspE genes is linked to cell adhesion, measured as FimH-mediated agglutination of yeast cells. While the UspC and UspE proteins promote motility at the expense of adhesion, the UspF and UspG proteins exhibit the exact opposite effects. We suggest that the Usp proteins have evolved different physiological functions that reprogram the cell towards defense and escape during cellular stress.
Figures
FIG. 1.
Effect of oxidative stress on growth of usp mutants. Fresh LB medium was inoculated with overnight culture of the different strains. In exponential phase, the precultures were diluted and split in two cultures, one of which was treated with 15 μM PMS. Open symbols, growth without PMS; filled symbols, growth in the presence of PMS; squares, wild-type strain MG1655; triangles, usp mutant strain. The mutant tested is shown on each panel, and arrows indicate the time of PMS addition. All experiments were performed at least three times to assess reproducibility. The figure presents the results of one typical experiment.
FIG. 2.
Effect of streptonigrin on growth of usp mutants. Fresh LB medium was inoculated with overnight culture of the different strains. In exponential phase, the precultures were diluted and split in two cultures, one of which was treated with streptonigrin (1 μg ml−1). Open symbols, growth without streptonigrin; filled symbols, growth in the presence of streptonigrin; squares, wild-type strain MG1655; triangles, usp mutant strain. The mutant tested is shown on each panel, and arrows indicate the time of streptonigrin addition. All experiments were performed at least three times to assess reproducibility. The figure presents the results of one typical experiment.
FIG. 3.
Effect of usp deletions on cellular autoaggregation. (A) Overnight cultures were vigorously shaken and subsequently incubated statically at room temperature. Upper panel: an aliquot of each culture was taken at the beginning of the assay for microscopic observation. Images were captured with a magnification of ×1,000. Lower panel: cell settling state of each strain after 1 h. (B) Ag43 production. For each strain, the equivalent of 0.2 OD600 unit of an overnight culture was analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and immunodetection using a polyclonal rabbit antiserum raised against the α domain of Ag43.
FIG. 4.
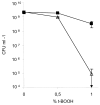
Survival of stationary-phase cells after challenge with the oxidative agent t-BOOH. Cells from overnight culture were incubated for 1 h at 37°C with different concentrations of t-BOOH. The survival is expressed as the number of CFU per ml on the LB plate after t-BOOH exposure. Symbols: ▪, wild-type strain MG1655; ▵, LN82 (uspE). Each point represents the average value calculated from the results of at least three experiments, and error bars indicate the calculated standard deviation.
FIG. 5.
(A) Effect of usp deletions on cell motility. Low-agar-concentration (0.3%) plates were inoculated with each strain as indicated. The halo of growth was observed after 48 h incubation at room temperature. (B) Detection of flagella by scanning electron microscopy. The observed strain is indicated in each panel. The black bar on each panel represents 2 μm.
FIG. 6.
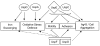
Role of the 6 E. coli Usps in oxidative stress defense, iron metabolism, and cell surface properties. The name of each Usp is surrounded by a shape symbolizing the class it belongs to: a circle for class I, a square for class II, and a diamond for class III and IV. An arrow represents a positive effect of the Usp protein in a specific function, whereas a T shape signifies a negative effect. Major and minor effects of the Usps in the different functions are represented by solid and dashed lines, respectively. The brackets indicate that both of the included proteins are involved in the indicated process. For example, UspC and UspE both affect motility positively and adhesion negatively.
Comment in
- Universal stress proteins in Escherichia coli.
Siegele DA. Siegele DA. J Bacteriol. 2005 Sep;187(18):6253-4. doi: 10.1128/JB.187.18.6253-6254.2005. J Bacteriol. 2005. PMID: 16159755 Free PMC article. No abstract available.
Similar articles
- The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage.
Gustavsson N, Diez A, Nyström T. Gustavsson N, et al. Mol Microbiol. 2002 Jan;43(1):107-17. doi: 10.1046/j.1365-2958.2002.02720.x. Mol Microbiol. 2002. PMID: 11849540 - Metabolic control of the Escherichia coli universal stress protein response through fructose-6-phosphate.
Persson O, Valadi A, Nyström T, Farewell A. Persson O, et al. Mol Microbiol. 2007 Aug;65(4):968-78. doi: 10.1111/j.1365-2958.2007.05838.x. Epub 2007 Jul 19. Mol Microbiol. 2007. PMID: 17640273 - The bacterial universal stress protein: function and regulation.
Kvint K, Nachin L, Diez A, Nyström T. Kvint K, et al. Curr Opin Microbiol. 2003 Apr;6(2):140-5. doi: 10.1016/s1369-5274(03)00025-0. Curr Opin Microbiol. 2003. PMID: 12732303 Review. - The universal stress protein UspC scaffolds the KdpD/KdpE signaling cascade of Escherichia coli under salt stress.
Heermann R, Weber A, Mayer B, Ott M, Hauser E, Gabriel G, Pirch T, Jung K. Heermann R, et al. J Mol Biol. 2009 Feb 13;386(1):134-48. doi: 10.1016/j.jmb.2008.12.007. Epub 2008 Dec 11. J Mol Biol. 2009. PMID: 19101563 - The Escherichia coli groE chaperonins.
Georgopoulos C, Ang D. Georgopoulos C, et al. Semin Cell Biol. 1990 Feb;1(1):19-25. Semin Cell Biol. 1990. PMID: 1983267 Review.
Cited by
- Antidepressants promote the spread of antibiotic resistance via horizontally conjugative gene transfer.
Ding P, Lu J, Wang Y, Schembri MA, Guo J. Ding P, et al. Environ Microbiol. 2022 Nov;24(11):5261-5276. doi: 10.1111/1462-2920.16165. Epub 2022 Aug 25. Environ Microbiol. 2022. PMID: 36054646 Free PMC article. - Stress responses in the opportunistic pathogen Acinetobacter baumannii.
Fiester SE, Actis LA. Fiester SE, et al. Future Microbiol. 2013 Mar;8(3):353-65. doi: 10.2217/fmb.12.150. Future Microbiol. 2013. PMID: 23464372 Free PMC article. Review. - SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato.
Loukehaich R, Wang T, Ouyang B, Ziaf K, Li H, Zhang J, Lu Y, Ye Z. Loukehaich R, et al. J Exp Bot. 2012 Sep;63(15):5593-606. doi: 10.1093/jxb/ers220. Epub 2012 Aug 21. J Exp Bot. 2012. PMID: 22915741 Free PMC article. - The α-helical regions of KERP1 are important in Entamoeba histolytica adherence to human cells.
Perdomo D, Baron B, Rojo-Domínguez A, Raynal B, England P, Guillén N. Perdomo D, et al. Sci Rep. 2013;3:1171. doi: 10.1038/srep01171. Epub 2013 Jan 30. Sci Rep. 2013. PMID: 23378906 Free PMC article. - Host genetic selection for cold tolerance shapes microbiome composition and modulates its response to temperature.
Kokou F, Sasson G, Nitzan T, Doron-Faigenboim A, Harpaz S, Cnaani A, Mizrahi I. Kokou F, et al. Elife. 2018 Nov 20;7:e36398. doi: 10.7554/eLife.36398. Elife. 2018. PMID: 30454554 Free PMC article.
References
- Aravind, L., V. Anantharaman, and E. V. Koonin. 2002. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins 48:1-14. - PubMed
- Bochkavera, E. S., A. S. Girshovich, and E. Bibi. 2002. Identification and characterization of the Escherichia coli stress protein UP12, a putative in vivo substrate of GroEL. Eur. J. Biochem. 269:3032-3040. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases