Whole-genome validation of high-information-content fingerprinting - PubMed (original) (raw)
Whole-genome validation of high-information-content fingerprinting
William M Nelson et al. Plant Physiol. 2005 Sep.
Abstract
Fluorescent-based high-information-content fingerprinting (HICF) techniques have recently been developed for physical mapping. These techniques make use of automated capillary DNA sequencing instruments to enable both high-resolution and high-throughput fingerprinting. In this article, we report the construction of a whole-genome HICF FPC map for maize (Zea mays subsp. mays cv B73), using a variant of HICF in which a type IIS restriction enzyme is used to generate the fluorescently labeled fragments. The HICF maize map was constructed from the same three maize bacterial artificial chromosome libraries as previously used for the whole-genome agarose FPC map, providing a unique opportunity for direct comparison of the agarose and HICF methods; as a result, it was found that HICF has substantially greater sensitivity in forming contigs. An improved assembly procedure is also described that uses automatic end-merging of contigs to reduce the effects of contamination and repetitive bands. Several new features in FPC v7.2 are presented, including shared-memory multiprocessing, which allows dramatically faster assemblies, and automatic end-merging, which permits more accurate assemblies. It is further shown that sequenced clones may be digested in silico and located accurately on the HICF assembly, despite size deviations that prevent the precise prediction of experimental fingerprints. Finally, repetitive bands are isolated, and their effect on the assembly is studied.
Figures
Figure 1.
Three-color fluorescent fingerprinting based on the type IIS restriction enzyme _Ear_I and 4-cutter _Taq_I. Shown are the (A) restriction enzyme cutting patterns, (B) the assignments of dye label to ddNTP and the ROX (−250 bp) standard that was used, (C) an example of enzyme cutting and labeling, and (D) a sample HICF trace (ZMMBBc0519B22) displayed in Genescan.
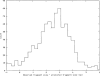
Figure 2.
Distribution of fragment sizing error from ABI 3700 sequencers, as determined by comparing in silico size predictions with observed fragment sizes for 22 sequenced maize clones (see text). The data are plotted as a histogram with a bin size of 0.25 bp.
Similar articles
- LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes.
Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. Frenkel Z, et al. BMC Bioinformatics. 2010 Nov 30;11:584. doi: 10.1186/1471-2105-11-584. BMC Bioinformatics. 2010. PMID: 21118513 Free PMC article. - A BAC pooling strategy combined with PCR-based screenings in a large, highly repetitive genome enables integration of the maize genetic and physical maps.
Yim YS, Moak P, Sanchez-Villeda H, Musket TA, Close P, Klein PE, Mullet JE, McMullen MD, Fang Z, Schaeffer ML, Gardiner JM, Coe EH Jr, Davis GL. Yim YS, et al. BMC Genomics. 2007 Feb 9;8:47. doi: 10.1186/1471-2164-8-47. BMC Genomics. 2007. PMID: 17291341 Free PMC article. - Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization.
Yim YS, Davis GL, Duru NA, Musket TA, Linton EW, Messing JW, McMullen MD, Soderlund CA, Polacco ML, Gardiner JM, Coe EH Jr. Yim YS, et al. Plant Physiol. 2002 Dec;130(4):1686-96. doi: 10.1104/pp.013474. Plant Physiol. 2002. PMID: 12481051 Free PMC article. - Organization and variability of the maize genome.
Messing J, Dooner HK. Messing J, et al. Curr Opin Plant Biol. 2006 Apr;9(2):157-63. doi: 10.1016/j.pbi.2006.01.009. Epub 2006 Feb 3. Curr Opin Plant Biol. 2006. PMID: 16459130 Review. - The Maize Genome Sequencing Project.
Chandler VL, Brendel V. Chandler VL, et al. Plant Physiol. 2002 Dec;130(4):1594-7. doi: 10.1104/pp.015594. Plant Physiol. 2002. PMID: 12481042 Free PMC article. Review. No abstract available.
Cited by
- LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes.
Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. Frenkel Z, et al. BMC Bioinformatics. 2010 Nov 30;11:584. doi: 10.1186/1471-2105-11-584. BMC Bioinformatics. 2010. PMID: 21118513 Free PMC article. - Recent advances in cotton genomics.
Zhang HB, Li Y, Wang B, Chee PW. Zhang HB, et al. Int J Plant Genomics. 2008;2008:742304. doi: 10.1155/2008/742304. Int J Plant Genomics. 2008. PMID: 18288253 Free PMC article. - Feasibility of physical map construction from fingerprinted bacterial artificial chromosome libraries of polyploid plant species.
Luo MC, Ma Y, You FM, Anderson OD, Kopecký D, Simková H, Safár J, Dolezel J, Gill B, McGuire PE, Dvorak J. Luo MC, et al. BMC Genomics. 2010 Feb 19;11:122. doi: 10.1186/1471-2164-11-122. BMC Genomics. 2010. PMID: 20170511 Free PMC article. - A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor.
Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen CM, Zhang Y, McGuire PE, Pasternak S, Stein JC, Ware D, Kramer M, McCombie WR, Kianian SF, Martis MM, Mayer KF, Sehgal SK, Li W, Gill BS, Bevan MW, Simková H, Dolezel J, Weining S, Lazo GR, Anderson OD, Dvorak J. Luo MC, et al. Proc Natl Acad Sci U S A. 2013 May 7;110(19):7940-5. doi: 10.1073/pnas.1219082110. Epub 2013 Apr 22. Proc Natl Acad Sci U S A. 2013. PMID: 23610408 Free PMC article.
References
- Bennett MD, Laurie DA (1995) Chromosome size in maize and sorghum using EM serial section reconstructed nuclei. Maydica 40: 199–204
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources