A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation - PubMed (original) (raw)
A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation
Shomyseh Sanjabi et al. Genes Dev. 2005.
Abstract
The NF-kappaB family members p65 (RelA) and c-Rel recognize similar DNA sequences, yet the phenotypes of mutant mice suggest that these proteins regulate distinct sets of genes. Here we demonstrate that 46 unique residues within an 86-residue segment of the Rel homology region (RHR) of c-Rel are responsible for the c-Rel requirement for Il12b gene induction by lipopolysaccharide in bone marrow-derived macrophages. These same residues were responsible for the c-Rel requirement for Il12a induction in dendritic cells, and in both instances, no evidence of c-Rel-specific coactivator interactions was found. Although the residues of c-Rel and p65 that contact specific bases and the DNA backbone within nuclear factor-kappaB (NF-kappaB) recognition sequences are identical, homodimers of c-Rel and of a chimeric p65 protein containing the critical c-Rel residues bound with high affinity to a broader range of NF-kappaB recognition sequences than did wild-type p65 homodimers. These results demonstrate that the unique functions of closely related transcription factor family members can be dictated by differences in the range of DNA sequences recognized at high affinity, despite having similar binding site consensus sequences and DNA contact residues.
Figures
Figure 1.
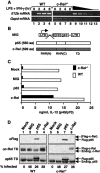
Retroviral expression of c-Rel in c-Rel-/- macrophages rescues Il12b expression. (A) RT-PCR was performed using RNA from wild-type and c-Rel-/- bone marrow-derived macrophages at the indicated time points after activation with LPS (10 μg/mL) and IFN-γ (10 U/mL). Twofold titrations of wild-type RNA (lanes _9_-12) and a negative control (lane 13) were included. (B) Diagrams of the MIG vector and murine p65 and c-Rel cDNAs are shown. The N-terminal [RHR(N)] and C-terminal [RHR(C)] portions of the RHR and transactivation domain (TD) are depicted. (C) Secreted IL-12 (p40/p70) protein levels observed in LPS/IFN-γ-activated wild-type and c-Rel-/- macrophages are shown. Cells were left untransduced (mock) or were transduced with the MIG virus or with viruses expressing p65 or c-Rel. (D) Protein levels in untransduced and transduced macrophages were monitored by Western blot, using antibodies directed against the Flag epitope or transactivation domains of c-Rel or p65. The percentage of cells transduced by each retroviral preparation was determined by flow cytometry analysis of GFP fluorescence.
Figure 2.
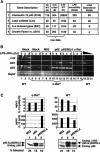
The N-terminal portion of the c-Rel RHR is necessary for Il12b induction in macrophages. (A) Primary amino acid sequences of mouse c-Rel and p65 RHRs were compared. Arrows indicate the boundaries between the N-terminal RHR (N), the C-terminal RHR (C), the transactivation domain (TD), and the first nonhomologous six and 18 residues of c-Rel and p65, respectively (ΔN; residues not shown). The N-terminal portion of the RHR was further divided into four regions labeled 1-4 (separated by brackets). The conserved DNA-contacting lysines in region 3 are underlined. The eight residues that were introduced in the modified proteins described in Supplementary Figure 5 are shown above the main sequence. (B) IL-12 (p40/p70) protein levels were measured after transduction of c-Rel-/- or wild-type macrophages with viruses expressing the indicated chimeras, and 24 h after stimulation with LPS + IFN-γ. Regions in white are from p65, and shaded regions are from c-Rel. The chimeras were named according to the following code: p65/domain X (N, C, and/or TD) from c-Rel. (C) Protein expression and the percentage of transduced cells in each sample were monitored as described in the legend to Figure 1D. The molecular weight of each protein is also shown.
Figure 3.
A p65/c-Rel chimera containing 46 residues from the c-Rel RHR activates Il12b expression in c-Rel-/- macrophages. (A) The wild-type and p65/c-Rel chimeras analyzed in this experiment are shown. (B) c-Rel-/- macrophages were transduced with various concentrations (twofold titrations) of titered viruses expressing each of the proteins depicted in A. (Rows 1, 2) Flow cytometry monitoring GFP fluorescence was used to determine the geometric mean fluorescence within each population, as well as the percentage of infected cells. (Row 3) IL-12 p40/p70 protein was monitored by ELISA. Western blots with antibodies against the Flag epitope, c-Rel TD, and p65 TD were used to monitor protein expression. An unknown protein recognized nonspecifically by the Flag antibody served as a control. (C) IL-12 p40/p70 protein (top) and Il12b mRNA (bottom) were analyzed by ELISA and RT-PCR, respectively, after transduction of c-Rel-/- macrophages and activation with LPS/IFN-γ for 8 h. Gapd mRNA was monitored as a control. (D) A structural model of the c-Rel RHR homodimer bound to the CD28 response element from the IL-2 promoter is shown (Huang et al. 2001). The DNA (white), region N3,4 of the c-Rel RHR (red), and regions N1,2 and C (green) are highlighted.
Figure 4.
Activation of other c-Rel-dependent genes by retroviral transduction. (A) Affymetrix microarrays were used to monitor gene expression in unstimulated wild-type macrophages (bone marrow-derived) and in stimulated wild-type and c-Rel-/- macrophages (10 μg/mL LPS + 10 U/mL IFN-γ for 8 h). Of 4849 expressed genes, 293 were up-regulated twofold or more by LPS + IFN-γ in wild-type cells, and 65 exhibited reduced expression (twofold or more) in stimulated c-Rel-/- cells versus stimulated wild-type cells. However, only four LPS/IFN-γ-stimulated genes exhibited c-Rel dependence. The raw data represent the normalized expression level from the Affymetrix arrays. (B) Wild-type and c-Rel-/- bone marrow-derived macrophages were mock-treated or transduced with the indicated viruses. After LPS/IFN-γ activation for 0, 3, or 8 h, total RNA was isolated and analyzed by semiquantitative RT-PCR. A two-fold titration of the wild-type 8-h-activated sample was used to monitor linearity of the RT-PCR assay. (C) Bone marrow precursors were differentiated into dendritic cells and were transduced with the MIG, p65, and p65/N3,4 viruses, followed by stimulation with LPS (1 μg/mL). Total IL-12 p40 concentrations (p40/p70, top graph) were compared with IL-12 p70 concentrations (bottom graph) as a strategy for monitoring the c-Rel requirement for IL-12 p35 expression (Grumont et al. 2001). Results from unstimulated and stimulated cells are shown (open and filled bars, respectively). Protein expression was monitored by Western blot, using a mixture of antibodies directed against the p65 TD and c-Rel TD.
Figure 5.
Enhanced binding of c-Rel and p65/N3,4 homodimers to NF-κB recognition sequences. (A) The sequences of EMSA probes containing the Il12b and Igk NF-κB recognition sequences are shown. (B) HEK 293T cells were transfected with plasmids expressing flag-tagged versions of c-Rel, p65, and p65/N3,4, with a small amount of a plasmid expressing murine p50. After extract preparation, EMSA experiments were performed using the Il12b (lanes _1_-12) and Igk (lanes _19_-30) probes, with different concentrations (twofold titrations) of each extract. (Lanes _13_-18) The composition of each complex was determined by supershift analysis using Flag and p50 antibodies. (Middle panel) A Western blot performed with Flag antibodies was used to monitor protein expression.
Figure 6.
Analysis of NF-κB recognition site mutants in a stable transfection assay. (A) At the top, the mouse Il12b promoter sequence from -154 to -104 was compared with the human Il12b promoter sequence from -138 to -88. The similarity between the distal NF-κB recognition sequence and the CD28 response element is shown, as are the sequences of the M1, M2, and M1,2 mutants analyzed. J774 cells were transfected with GFP reporter plasmids containing the Il12b promoter alone (_Il12b_P), Il12b promoter with an upstream Il12b enhancer (_Il12b_EP), or mutant Il12b promoters with the upstream enhancer (M1, M2, and M1,2). Cell clones were stimulated with LPS and GFP reporter activity monitored by flow cytometry. Each dot represents the mean fluorescence observed with a different cell clone, after subtraction of the mean fluorescence observed with the unstimulated cells of the same clone. (B) Binding of c-Rel and p65 to the endogenous Il12b promoter in bone marrow-derived macrophages stimulated with LPS + IFN-γ was monitored by ChIP. Primers that amplify the IκBα and MIP-2 promoters, which are also induced by LPS, were included as positive controls. Primers that amplify the IL-2 promoter were included as a negative control. Control genomic DNA samples at the right demonstrate the relative amplification efficiencies of the different primer pairs, and input samples at the bottom demonstrate that similar amounts of chromatin from each time point were analyzed.
Figure 7.
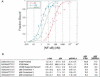
Relative affinities of the c-Rel, p65, and p65/N3,4 RHRs for NF-κB recognition sequences. (A) Compiled results are shown from three independent EMSA titrations comparing the binding of p65, p65/N3,4, and c-Rel to the Il12b proximal recognition site. Results were normalized for the percentage of active protein within each purified protein sample. The _Y_-axis shows the fraction of probe molecules bound by protein, and the _X_-axis shows the concentration of NF-κB homodimer added to the reaction. The plot was created by the Kaleidograph program, using a nonlinear least squares analysis as described by Cowley and Graves (2000). (B) Kd values (nanomolar, with standard deviations) are shown for recombinant NF-κB RHR homodimers bound to probes containing the indicated recognition sequences. The results were derived from three independent EMSA titrations performed with each probe and each RHR homodimer. Kd values were determined as described (Cowley and Graves 2000). At the right, Kd ratios for p65 versus c-Rel and for p65 versus p65/N3,4 are shown.
Similar articles
- Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Chen FE, Huang DB, Chen YQ, Ghosh G. Chen FE, et al. Nature. 1998 Jan 22;391(6665):410-3. doi: 10.1038/34956. Nature. 1998. PMID: 9450761 - X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2.
Huang DB, Chen YQ, Ruetsche M, Phelps CB, Ghosh G. Huang DB, et al. Structure. 2001 Aug;9(8):669-78. doi: 10.1016/s0969-2126(01)00635-9. Structure. 2001. PMID: 11587641 - The structure of the NF-kappa B p50:DNA-complex: a starting point for analyzing the Rel family.
Müller CW, Harrison SC. Müller CW, et al. FEBS Lett. 1995 Aug 1;369(1):113-7. doi: 10.1016/0014-5793(95)00541-g. FEBS Lett. 1995. PMID: 7641872 Review. - Rel/NF-kappaB transcription factors: key mediators of B-cell activation.
Gugasyan R, Grumont R, Grossmann M, Nakamura Y, Pohl T, Nesic D, Gerondakis S. Gugasyan R, et al. Immunol Rev. 2000 Aug;176:134-40. doi: 10.1034/j.1600-065x.2000.00615.x. Immunol Rev. 2000. PMID: 11043773 Review.
Cited by
- IL-10 constrains sphingolipid metabolism to limit inflammation.
York AG, Skadow MH, Oh J, Qu R, Zhou QD, Hsieh WY, Mowel WK, Brewer JR, Kaffe E, Williams KJ, Kluger Y, Smale ST, Crawford JM, Bensinger SJ, Flavell RA. York AG, et al. Nature. 2024 Mar;627(8004):628-635. doi: 10.1038/s41586-024-07098-5. Epub 2024 Feb 21. Nature. 2024. PMID: 38383790 Free PMC article. - Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses.
Liu PT, Schenk M, Walker VP, Dempsey PW, Kanchanapoomi M, Wheelwright M, Vazirnia A, Zhang X, Steinmeyer A, Zügel U, Hollis BW, Cheng G, Modlin RL. Liu PT, et al. PLoS One. 2009 Jun 5;4(6):e5810. doi: 10.1371/journal.pone.0005810. PLoS One. 2009. PMID: 19503839 Free PMC article. - RNAi-mediated c-Rel silencing leads to apoptosis of B cell tumor cells and suppresses antigenic immune response in vivo.
Tian W, Liou HC. Tian W, et al. PLoS One. 2009;4(4):e5028. doi: 10.1371/journal.pone.0005028. Epub 2009 Apr 6. PLoS One. 2009. PMID: 19347041 Free PMC article. - Transcriptional regulation of Acsl1 by CHREBP and NF-kappa B in macrophages during hyperglycemia and inflammation.
Thevkar-Nagesh P, Habault J, Voisin M, Ruff SE, Ha S, Ruoff R, Chen X, Rawal S, Zahr T, Szabo G, Rogatsky I, Fisher EA, Garabedian MJ. Thevkar-Nagesh P, et al. PLoS One. 2022 Sep 2;17(9):e0272986. doi: 10.1371/journal.pone.0272986. eCollection 2022. PLoS One. 2022. PMID: 36054206 Free PMC article. - c-Rel plays a key role in deficient activation of B cells from a non-X-linked hyper-IgM patient.
Lu KT, Sinquett FL, Dryer RL, Song C, Covey LR. Lu KT, et al. Blood. 2006 Dec 1;108(12):3769-76. doi: 10.1182/blood-2006-03-008839. Epub 2006 Aug 8. Blood. 2006. PMID: 16896156 Free PMC article. Clinical Trial.
References
- Attar R.M., Caamaño, J., Carrasco, D., Iotsova, V., Ishikawa, H., Ryseck, R.-P., Weih, F., and Bravo, R. 1997. Genetic approaches to study Rel/NF-κB/IκB function in mice. Semin. Cancer Biol. 8: 93-101. - PubMed
- Beg A.A. and Baltimore, D. 1996. An essential role for NF-κBin preventing TNF-α-induced cell death. Science 274: 782-784. - PubMed
- Bonizzi G. and Karin, M. 2004. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25: 280-288. - PubMed
- Carey M. and Smale, S.T. 2000. Transcriptional regulation in eukaryotes: Concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Carlsson P. and Mahlapuu, M. 2002. Forkhead transcription factors: Key players in development and metabolism. Dev. Biol. 250: 1-23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials