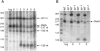Small untranslated RNA antitoxin in Bacillus subtilis - PubMed (original) (raw)
Small untranslated RNA antitoxin in Bacillus subtilis
Jessica M Silvaggi et al. J Bacteriol. 2005 Oct.
Abstract
Toxin-antitoxin (TA) modules are pairs of genes in which one member encodes a toxin that is neutralized or whose synthesis is prevented by the action of the product of the second gene, an antitoxin, which is either protein or RNA. We now report the identification of a TA module in the chromosome of Bacillus subtilis in which the antitoxin is an antisense RNA. The antitoxin, which is called RatA (for RNA antitoxin A), is a small (222 nucleotides), untranslated RNA that blocks the accumulation of the mRNA for a toxic peptide TxpA (for toxic peptide A; formerly YqdB). The txpA and ratA genes are in convergent orientation and overlap by ca. 75 nucleotides, such that the 3' region of ratA is complementary to the 3' region of txpA. Deletion of ratA led to increased levels of txpA mRNA and lysis of the cells. Overexpression of txpA also caused cell lysis and death, a phenotype that was prevented by simultaneous overexpression of ratA. We propose that the ratA transcript is an antisense RNA that anneals to the 3' end of the txpA mRNA, thereby triggering its degradation.
Figures
FIG. 1.
txpA-yqbM intergenic region. Depicted are both DNA strands of the intergenic region and flanking DNA and the orientation and location of ratA and protein-coding genes adjacent to the intergenic region. Transcriptional start sites are indicated by +1. A predicted stem-loop terminator is indicated at the end of ratA. Also depicted is the riboprobe that was used to detect txpA mRNA. Shown at the top are the boundaries of the intergenic region and segments of DNA deleted in the igr-1 and igr-2 mutations. Shown at the bottom are segments of DNA used in the complementation analysis with a “+” indicating complementation by the indicated DNA segment and a “-” indicating the absence of complementation.
FIG. 2.
Detection of RatA by Northern blot analysis. Panel A shows the results with total RNA (5 μg) from wild-type cells grown in DS medium at 37°C and collected during the exponential phase of growth (log) and at hourly intervals during sporulation. Panel B shows the results with RNA from wild-type cells (WT) and _igr-1_-containing cells grown in DS medium and collected during the exponential phase of growth and at hours 0, 1, and 2 of sporulation. The blots of panels A and B were probed with a PCR-amplified, double-stranded DNA that corresponded to the full-length, intergenic region between txpA and yqbM. Shown on the left of the blots as molecular weight markers are 5′-end-labeled MspI fragments of pBR322 DNA that had been denatured.
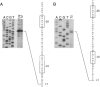
FIG. 3.
Mapping the 5′ ends of RatA and txpA mRNA. (A) Primer extension experiment with 20 μg of total RNA from wild-type cells grown in DS medium and collected during the exponential phase of growth (log) and at hour 2 of sporulation (T2). The RNAs were used as a template for extension with an end-labeled primer (oJS13) corresponding to ratA. A, C, G, and T indicate the dideoxy sequencing ladders generated with the same primer and terminated with the corresponding ddNTP. The corresponding cDNA sequence was determined from the DNA sequencing ladder. The “+1” indicates the 5′ end of the message. The boxes indicate putative −10 and −35 sequences. The dotted portion of the lower box indicates an extended −10 sequence. (B) Primer extension experiment similar to that of panel A except with an end-labeled primer corresponding to txpA (oJS466).
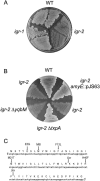
FIG. 4.
Complementation of the lysis phenotype caused by the absence of RatA. (A) LB agar plate streaked with the wild type (WT) and with mutants harboring the igr-1 or igr-2 mutations and grown at 37°C for 5 days. (B) LB agar plate streaked with the wild type (WT) and with mutants harboring igr-2 (JS178), igr-2 amyE::pJS63 (JS228), igr-2 Δ_yqbM_ (JS251), and igr-2 Δ_txpA_ (JS226) and grown at 37°C for 5 days. (C) Nucleotide and amino acid sequence for txpA. Indicated above the amino acid sequence are alterations due to suppressor mutations that arose in transformants generated by transformation with constructs containing deletion/insertion mutations of the _txpA_-yqbM intergenic region.
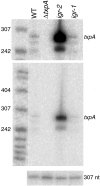
FIG. 5.
Visualization of txpA mRNA by Northern blot analysis. Total RNA (5 μg) was isolated from wild-type cells (WT) and from cells containing Δ_txpA_ (JS191), igr-2 (JS178), or igr-1 (JS94) that had been grown in DS medium at 37°C and collected during late exponential growth. The txpA transcript was detected by using the riboprobe depicted in Fig. 1, which was complementary to the 3′ end of txpA, representing the 75-nt region of overlap with RatA. The upper panel represents a threefold longer exposure of the middle panel for better visualization of the low level of txpA mRNA in the wild type. The bottom panel is a loading control probed with a double-stranded probe for an unrelated RNA of 307 nt. The molecular weight markers are 5′-end-labeled MspI fragments of pBR322 DNA that had been denatured.
FIG. 6.
Induction of txpA in the absence of RatA causes cell death. Panels A and C show OD measurements, and panels B and D show measurements of CFU. The experiment was carried out with a strain (JS348) in which ratA was under the control of a xylose-inducible promoter and txpA was under the control of an IPTG-inducible promoter. Panels A and B show the results for an experiment in which cells at the exponential phase of growth in LB medium at 37°C were split into two cultures, one of which was supplemented with 100 μM of IPTG (•) and the other was not induced (○). Time represents minutes after the addition of inducer. Panels C and D show the results for an experiment in which cells at the exponential phase of growth in LB medium were split into three cultures. One culture was treated simultaneously with 130 mM xylose and 100 μM IPTG (▴) at time zero. The second culture was treated with xylose at time zero and then with IPTG at 60 min as indicated by the arrow (□). The third culture was treated with IPTG at time zero and then with xylose at 60 min (▪).
Similar articles
- Type I toxin-antitoxin systems in Bacillus subtilis.
Durand S, Jahn N, Condon C, Brantl S. Durand S, et al. RNA Biol. 2012 Dec;9(12):1491-7. doi: 10.4161/rna.22358. Epub 2012 Oct 11. RNA Biol. 2012. PMID: 23059907 Review. - Regulatory crosstalk between type I and type II toxin-antitoxin systems in the human pathogen Enterococcus faecalis.
Wessner F, Lacoux C, Goeders N, Fouquier d'Hérouel A, Matos R, Serror P, Van Melderen L, Repoila F. Wessner F, et al. RNA Biol. 2015;12(10):1099-108. doi: 10.1080/15476286.2015.1084465. Epub 2015 Aug 25. RNA Biol. 2015. PMID: 26305399 Free PMC article. - BsrG/SR4 from Bacillus subtilis--the first temperature-dependent type I toxin-antitoxin system.
Jahn N, Preis H, Wiedemann C, Brantl S. Jahn N, et al. Mol Microbiol. 2012 Feb;83(3):579-98. doi: 10.1111/j.1365-2958.2011.07952.x. Epub 2012 Jan 9. Mol Microbiol. 2012. PMID: 22229825 - Characterization of a Streptococcus mutans intergenic region containing a small toxic peptide and its cis-encoded antisense small RNA antitoxin.
Koyanagi S, Lévesque CM. Koyanagi S, et al. PLoS One. 2013;8(1):e54291. doi: 10.1371/journal.pone.0054291. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326602 Free PMC article. - sRNAs in bacterial type I and type III toxin-antitoxin systems.
Brantl S, Jahn N. Brantl S, et al. FEMS Microbiol Rev. 2015 May;39(3):413-27. doi: 10.1093/femsre/fuv003. Epub 2015 Mar 25. FEMS Microbiol Rev. 2015. PMID: 25808661 Review.
Cited by
- A genetic selection reveals functional metastable structures embedded in a toxin-encoding mRNA.
Masachis S, Tourasse NJ, Lays C, Faucher M, Chabas S, Iost I, Darfeuille F. Masachis S, et al. Elife. 2019 Aug 14;8:e47549. doi: 10.7554/eLife.47549. Elife. 2019. PMID: 31411564 Free PMC article. - AU-Rich Long 3' Untranslated Region Regulates Gene Expression in Bacteria.
Zhao JP, Zhu H, Guo XP, Sun YC. Zhao JP, et al. Front Microbiol. 2018 Dec 12;9:3080. doi: 10.3389/fmicb.2018.03080. eCollection 2018. Front Microbiol. 2018. PMID: 30619162 Free PMC article. - Agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern.
Rieu A, Weidmann S, Garmyn D, Piveteau P, Guzzo J. Rieu A, et al. Appl Environ Microbiol. 2007 Oct;73(19):6125-33. doi: 10.1128/AEM.00608-07. Epub 2007 Aug 3. Appl Environ Microbiol. 2007. PMID: 17675424 Free PMC article. - Toxin⁻Antitoxin Systems in Bacillus subtilis.
Brantl S, Müller P. Brantl S, et al. Toxins (Basel). 2019 May 9;11(5):262. doi: 10.3390/toxins11050262. Toxins (Basel). 2019. PMID: 31075979 Free PMC article. Review. - Bacillus subtilis Type I antitoxin SR6 Promotes Degradation of Toxin yonT mRNA and Is Required to Prevent Toxic yoyJ Overexpression.
Reif C, Löser C, Brantl S. Reif C, et al. Toxins (Basel). 2018 Feb 7;10(2):74. doi: 10.3390/toxins10020074. Toxins (Basel). 2018. PMID: 29414903 Free PMC article.
References
- Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. - PubMed
- Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. - PubMed
- Bernhardt, T. G., I. N. Wang, D. K. Struck, and R. Young. 2002. Breaking free: “protein antibiotics” and phage lysis. Res. Microbiol. 153:493-501. - PubMed
- Brantl, S. 2002. Antisense RNAs in plasmids: control of replication and maintenance. Plasmid 48:165-173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases