Slip sliding away: load-dependence of velocity generated by skeletal muscle myosin molecules in the laser trap - PubMed (original) (raw)
Slip sliding away: load-dependence of velocity generated by skeletal muscle myosin molecules in the laser trap
Edward P Debold et al. Biophys J. 2005 Nov.
Abstract
Skeletal muscle's ability to shorten and lengthen against a load is a fundamental property, presumably reflecting the inherent load-dependence of the myosin molecular motor. Here we report the velocity of a single actin filament translocated by a mini-ensemble of skeletal myosin approximately 8 heads under constant loads up to 15 pN in a laser trap assay. Actin filament velocity decreased with increasing load hyberbolically, with unloaded velocity and stall force differing by a factor of 2 with [ATP] (30 vs. 100 muM). Analysis of actin filament movement revealed that forward motion was punctuated with rapid backward 60-nm slips, with the slip frequency increasing with resistive load. At stall force, myosin-generated forward movement was balanced by backward slips, whereas at loads greater than stall, myosin could no longer sustain forward motion, resulting in negative velocities as in eccentric contractions of whole muscle. Thus, the force-velocity relationship of muscle reflects both the inherent load-dependence of the actomyosin interaction and the balance between forward and reverse motion observed at the molecular level.
Figures
FIGURE 1
(A) F/V data at 30 _μ_M (red circles) and 100 _μ_M ATP (black triangles) as means ± SE. Hill F/V fit to positive loads only (lines; see text) at 30 _μ_M ATP (_F_o = 15 pN, a/_F_o = 0.29, b = 0.21 _μ_m/s) and 100 _μ_M ATP (_F_o = 6 pN, a/_F_o = 0.33, b = 0.53 _μ_m/s). (B) Only increasing load sequence shown with force levels above displacement trace. (C) Filtered actin displacement traces (at 100 _μ_M ATP) at different loads with their respective linear regressions used to determine actin filament velocity; 10–90 velocities at each load were averaged to generate F/V in panel A.
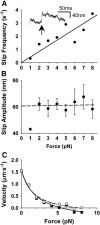
FIGURE 2
Characterization of slips at 100 _μ_M ATP (similar results obtained at 30 _μ_M ATP). (A) Slip frequency versus load (r = 0.84). Inset is a displacement record with a rapid reversal or “slip” in response to 5 pN. (B) Slip amplitude versus force, each point is the mean ± SE. Regression omits 1-pN data yielding 60-nm average amplitude. (C) F/V data with (▪, solid line) and without slips (○, dashed line) and fit with Hill F/V equation.
Similar articles
- The biochemical kinetics underlying actin movement generated by one and many skeletal muscle myosin molecules.
Baker JE, Brosseau C, Joel PB, Warshaw DM. Baker JE, et al. Biophys J. 2002 Apr;82(4):2134-47. doi: 10.1016/S0006-3495(02)75560-4. Biophys J. 2002. PMID: 11916869 Free PMC article. - Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap.
Guilford WH, Dupuis DE, Kennedy G, Wu J, Patlak JB, Warshaw DM. Guilford WH, et al. Biophys J. 1997 Mar;72(3):1006-21. doi: 10.1016/S0006-3495(97)78753-8. Biophys J. 1997. PMID: 9138552 Free PMC article. - A model of muscle contraction based on the Langevin equation with actomyosin potentials.
Tamura Y, Ito A, Saito M. Tamura Y, et al. Comput Methods Biomech Biomed Engin. 2017 Feb;20(3):273-283. doi: 10.1080/10255842.2016.1215440. Epub 2016 Jul 29. Comput Methods Biomech Biomed Engin. 2017. PMID: 27472485 - Actomyosin interaction in striated muscle.
Cooke R. Cooke R. Physiol Rev. 1997 Jul;77(3):671-97. doi: 10.1152/physrev.1997.77.3.671. Physiol Rev. 1997. PMID: 9234962 Review. - Mechanics and models of the myosin motor.
Huxley AF. Huxley AF. Philos Trans R Soc Lond B Biol Sci. 2000 Apr 29;355(1396):433-40. doi: 10.1098/rstb.2000.0584. Philos Trans R Soc Lond B Biol Sci. 2000. PMID: 10836496 Free PMC article. Review.
Cited by
- Isoforms Confer Characteristic Force Generation and Mechanosensation by Myosin II Filaments.
Stam S, Alberts J, Gardel ML, Munro E. Stam S, et al. Biophys J. 2015 Apr 21;108(8):1997-2006. doi: 10.1016/j.bpj.2015.03.030. Biophys J. 2015. PMID: 25902439 Free PMC article. - Cooperativity of myosin II motors in the non-regulated and regulated thin filaments investigated with high-speed AFM.
Matusovsky OS, Månsson A, Rassier DE. Matusovsky OS, et al. J Gen Physiol. 2023 Mar 6;155(3):e202213190. doi: 10.1085/jgp.202213190. Epub 2023 Jan 12. J Gen Physiol. 2023. PMID: 36633585 Free PMC article. - Mutation of a conserved glycine in the SH1-SH2 helix affects the load-dependent kinetics of myosin.
Kad NM, Patlak JB, Fagnant PM, Trybus KM, Warshaw DM. Kad NM, et al. Biophys J. 2007 Mar 1;92(5):1623-31. doi: 10.1529/biophysj.106.097618. Epub 2006 Dec 1. Biophys J. 2007. PMID: 17142278 Free PMC article. - Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function.
Schmitt JP, Debold EP, Ahmad F, Armstrong A, Frederico A, Conner DA, Mende U, Lohse MJ, Warshaw D, Seidman CE, Seidman JG. Schmitt JP, et al. Proc Natl Acad Sci U S A. 2006 Sep 26;103(39):14525-30. doi: 10.1073/pnas.0606383103. Epub 2006 Sep 18. Proc Natl Acad Sci U S A. 2006. PMID: 16983074 Free PMC article. - Smooth muscle heavy meromyosin phosphorylated on one of its two heads supports force and motion.
Walcott S, Fagnant PM, Trybus KM, Warshaw DM. Walcott S, et al. J Biol Chem. 2009 Jul 3;284(27):18244-51. doi: 10.1074/jbc.M109.003293. Epub 2009 May 6. J Biol Chem. 2009. PMID: 19419961 Free PMC article.
References
- Hill, A. V. 1938. The heat of shortening and the dynamic constants of muscle. Proc. R. Soc. Lond. B. Biol. Sci. 126:136–195.
- Huxley, A. F. 1957. Muscle structure and theories of contraction. Prog. Biophys. Biophys. Chem. 7:255–318. - PubMed
- Harris, D. E., and D. M. Warshaw. 1993. Smooth and skeletal muscle myosin both exhibit low duty cycles in vitro. J. Biol. Chem. 268:14764–14768. - PubMed
- Visscher, K., M. J. Schnitzer, and S. M. Block. 1999. Single kinesin molecules studied with a molecular force clamp. Nature. 400:184–189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources