Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein - PubMed (original) (raw)
Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein
Xiao-Yong Zhang et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The c-myc oncogene is among the most commonly overexpressed genes in human cancer. c-myc encodes a basic helix-loop-helix/leucine zipper (bHLH/LZ) transcription factor (c-MYC) that activates a cascade of downstream targets that ultimately mediate cellular transformation. Although a large number of genes are regulated by c-MYC, only a few have been functionally linked to c-MYC-mediated transformation. By expression profiling, the metastasis-associated protein 1 (MTA1) gene was identified here as a target of the c-MYC oncoprotein in primary human cells, a result confirmed in human cancer cells. MTA1 itself has been previously implicated in cellular transformation, in part through its ability to regulate the epithelial-to-mesenchymal transition and metastasis. MTA1 is a component of the Mi-2/nucleosome remodeling and deacetylating (NURD) complex that contains both histone deacetylase and nucleosome remodeling activity. The data reported here demonstrate that endogenous c-MYC binds to the genomic MTA1 locus and recruits transcriptional coactivators. Most importantly, short hairpin RNA (shRNA)-mediated knockdown of MTA1 blocks the ability of c-MYC to transform mammalian cells. These data implicate MTA1 and the Mi-2/NURD complex as one of the first downstream targets of c-MYC function that are essential for the transformation potential of c-MYC.
Figures
Fig. 1.
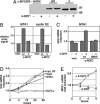
Identification of the breast cancer metastasis regulator MTA1 as a transcriptional target of the c-MYC oncoprotein. (A) Western blot analysis shows that the wild-type and ΔMbII mutant of the human c-MYC-estrogen receptor fusion protein were comparably expressed in stable pools of NHDF cells. (B) Microarray analysis was performed by using mRNA harvested from cells at 4 h after 4-OHT treatment. Array signals for MTA1 and cyclin D2 are shown for the 4-OHT treated groups. (C) qRT-PCR was performed to verify the induction of MTA1 transcription by c-MYC. (D) The kinetics of MTA1 transcript induction in NHDF cells after c-MYC/ER activation was determined. As controls, levels were also determined for the c-MYC target genes cyclin D2 and CAD. As a negative control, levels of ELF1a transcript levels were quantitated. Actin levels were used to normalize expression patterns between samples. (E) Regulation of MTA1 by c-MYC/ER in a murine B cell lymphoma was assessed. Cells from the tumor were explanted and adapted for growth in culture. These lymphoma cells were 4-OHT-treated for the times indicated, and MTA1 transcript levels were quantitated by qRT-PCR. Nontreated cells served as a negative control, and samples were normalized for actin mRNA expression.
Fig. 2.
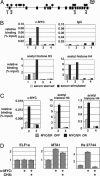
In vivo binding of c-MYC to the endogenous MTA1 locus in human cells. (A) The MTA1 genomic locus contains 16 exons dispersed over 60 kb (indicated by black boxes). Within the locus, several potential matches to the c-MYC consensus binding site were identified (filled circles). (B) Four of these sites (sites 1–4) were selected for direct examination of c-MYC binding by using the ChIP technique. For this analysis, c-MYC was induced in growth factor-deprived NHDFs by serum stimulation for 2 h. ChIP analysis was performed to detect DNA associated with the c-MYC protein or with acetylated histones H3 or H4. Nonimmune rabbit IgG was used in control ChIP experiments. (C) NHDF cells expressing the c-MYC/ER protein were treated with 4-OHT for 2 h and then subjected to ChIP analysis as above. (D) To test whether MTA1 is a direct or indirect target of c-MYC, c-MYC/ER-expressing NHDFs were used. After growth factor starvation, cells were pretreated with CHX for 30 min, and then 4-OHT was added in the presence of CHX for 4 h to activate c-MYC/ER. Cells were harvested and levels of MTA1 transcript were determined by qRT-PCR. As controls, levels of ELF1a and Hs27744 transcripts were also determined.
Fig. 3.
MTA1 levels are regulated by c-MYC in a murine tumor model and in human breast cancer cells. (A) A transplantable B cell lymphoma driven by c-MYC/ER was transferred to four naive mice. (B) After 9 days of 4-OHT treatment, mice developed aggressively growing lymphomas. At this point, two mice were continued on 4-OHT treatment (mice 1 and 2) whereas the remaining two mice were withdrawn from treatment for 4 days (mice 3 and 4). MTA1 mRNA levels in the tumors were quantitated. (C) Loss of endogenous c-MYC specifically inhibits MTA1 transcription in human cancer cells. Human breast (MCF7) and lung (H1299) cancer cells were transfected with c-MYC siRNA oligos (Dharmacon) or GFP siRNA as a control. Two days after transfection, c-MYC, MTA1, actin, and HDAC5 mRNA levels were determined by qRT-PCR. Transcript levels of these genes in GFP and c-MYC siRNA-treated cells are shown, by light and dark bars, respectively. (D) H1299 cells were lysed 5 days posttransfection with constructs encoding either a control (con.) or c-MYC shRNA vector. Protein lysates were analyzed by Western blotting for MTA1, c-MYC, or actin, as indicated. Western blot signals for MTA1 and c-MYC were quantitated and normalized to actin levels, and numbers indicate these normalized levels.
Fig. 4.
MTA1 expression is essential for c-MYC-induced growth in soft agar. (A) Rat1a fibroblasts expressing the c-MYC/ER protein were infected with a retrovirus encoding either of two distinct shRNAs targeting MTA1. In parallel, cells were infected with a retrovirus encoding an irrelevant shRNA as a control (con.). Seventy-two hours after infection with shRNA retroviruses, mRNA was harvested from Rat1a c-MYC/ER cells and analyzed for MTA1 transcript levels. (B) MTA1 protein levels were determined by Western blotting of cell lysates after knockdown in Rat1a c-MYC/ER cells. Blots were probed for tubulin as a control for protein loading. (C) After MTA1 knockdown, Rat1a c-MYC/ER cells were treated with 4-OHT to activate c-MYC and then analyzed for growth in soft agar. Mock-treated cells served as a control. Images show colony size at 7 days after c-MYC activation. (D) Low magnification images of soft agar assay wells.
Fig. 5.
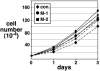
MTA1 expression is not essential for c-MYC-induced cell cycle progression. Rat1a cells expressing c-MYC/ER were infected with retroviral stocks encoding the two distinct MTA1 shRNA constructs or a control shRNA as indicated. After selection for infected cells, cells were plated in the presence (solid lines) or absence (dashed lines) of 4-OHT. Cell numbers were determined by direct counting of triplicate wells at each of the time points indicated.
Similar articles
- The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications.
Toh Y, Nicolson GL. Toh Y, et al. Clin Exp Metastasis. 2009;26(3):215-27. doi: 10.1007/s10585-008-9233-8. Epub 2008 Dec 31. Clin Exp Metastasis. 2009. PMID: 19116762 Review. - Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1.
Khaleque MA, Bharti A, Gong J, Gray PJ, Sachdev V, Ciocca DR, Stati A, Fanelli M, Calderwood SK. Khaleque MA, et al. Oncogene. 2008 Mar 20;27(13):1886-93. doi: 10.1038/sj.onc.1210834. Epub 2007 Oct 8. Oncogene. 2008. PMID: 17922035 - Identification of novel targets of MYC whose transcription requires the essential MbII domain.
Zhang XY, DeSalle LM, McMahon SB. Zhang XY, et al. Cell Cycle. 2006 Feb;5(3):238-41. doi: 10.4161/cc.5.3.2409. Epub 2006 Feb 1. Cell Cycle. 2006. PMID: 16434883 - MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene.
Molli PR, Singh RR, Lee SW, Kumar R. Molli PR, et al. Oncogene. 2008 Mar 27;27(14):1971-80. doi: 10.1038/sj.onc.1210839. Epub 2007 Oct 8. Oncogene. 2008. PMID: 17922032 Free PMC article. - Clinical implications of MTA proteins in human cancer.
Kaur E, Gupta S, Dutt S. Kaur E, et al. Cancer Metastasis Rev. 2014 Dec;33(4):1017-24. doi: 10.1007/s10555-014-9527-z. Cancer Metastasis Rev. 2014. PMID: 25374266 Review.
Cited by
- The Quest for Targets Executing MYC-Dependent Cell Transformation.
Hartl M. Hartl M. Front Oncol. 2016 Jun 2;6:132. doi: 10.3389/fonc.2016.00132. eCollection 2016. Front Oncol. 2016. PMID: 27313991 Free PMC article. Review. - Epigenetics of prostate cancer: beyond DNA methylation.
Schulz WA, Hatina J. Schulz WA, et al. J Cell Mol Med. 2006 Jan-Mar;10(1):100-25. doi: 10.1111/j.1582-4934.2006.tb00293.x. J Cell Mol Med. 2006. PMID: 16563224 Free PMC article. Review. - Role of MTA1 in cancer progression and metastasis.
Sen N, Gui B, Kumar R. Sen N, et al. Cancer Metastasis Rev. 2014 Dec;33(4):879-89. doi: 10.1007/s10555-014-9515-3. Cancer Metastasis Rev. 2014. PMID: 25344802 Free PMC article. Review. - MYCN and the epigenome.
He S, Liu Z, Oh DY, Thiele CJ. He S, et al. Front Oncol. 2013 Jan 25;3:1. doi: 10.3389/fonc.2013.00001. eCollection 2013. Front Oncol. 2013. PMID: 23373009 Free PMC article. - Chromatin Remodelers: From Function to Dysfunction.
Längst G, Manelyte L. Längst G, et al. Genes (Basel). 2015 Jun 12;6(2):299-324. doi: 10.3390/genes6020299. Genes (Basel). 2015. PMID: 26075616 Free PMC article. Review.
References
- Nesbit, C. E., Tersak, J. M. & Prochownik, E. V. (1999) Oncogene 18, 3004–3016. - PubMed
- Nilsson, J. A. & Cleveland, J. L. (2003) Oncogene 22, 9007–9021. - PubMed
- Cole, M. D. & McMahon, S. B. (1999) Oncogene 18, 2916–2924. - PubMed
- Lewis, B. C., Prescott, J. E., Campbell, S. E., Shim, H., Orlowski, R. Z. & Dang, C. V. (2000) Cancer Res. 60, 6178–6183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA090465/CA/NCI NIH HHS/United States
- CA102709/CA/NCI NIH HHS/United States
- R01 CA098172/CA/NCI NIH HHS/United States
- R21 CA097932/CA/NCI NIH HHS/United States
- R01 CA102709/CA/NCI NIH HHS/United States
- R01 CA090465/CA/NCI NIH HHS/United States
- CA097932/CA/NCI NIH HHS/United States
- CA098172/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous