Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A - PubMed (original) (raw)
Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A
Anna Scott et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The VPS4 AAA ATPases function both in endosomal vesicle formation and in the budding of many enveloped RNA viruses, including HIV-1. VPS4 proteins act by binding and catalyzing release of the membrane-associated ESCRT-III protein lattice, thereby allowing multiple rounds of protein sorting and vesicle formation. Here, we report the solution structure of the N-terminal VPS4A microtubule interacting and transport (MIT) domain and demonstrate that the VPS4A MIT domain binds the C-terminal half of the ESCRT-III protein, CHMP1B (Kd = 20 +/- 13 microM). The MIT domain forms an asymmetric three-helix bundle that resembles the first three helices in a tetratricopeptide repeat (TPR) motif. Unusual interhelical interactions are mediated by a series of conserved aromatic residues that form coiled-coil interactions between the second two helices and also pack against the conserved alanines that interdigitate between the first two helices. Mutational analyses revealed that a conserved leucine residue (Leu-64) on the third helix that would normally bind the fourth helix in an extended TPR is used to bind CHMP1B, raising the possibility that ESCRT-III proteins may bind by completing the TPR motif.
Figures
Fig. 1.
Sequences and features of VPS4A, CHMP1B, and MIT domains. (A) Schematic illustrations of human VPS4A (19) and CHMP1B. The predicted coiled-coil (CC) of CHMP1B was identified at a probability of 0.17 by using
multicoil
(41). (B) Sequence alignment of known MIT domains from eight human proteins (Upper) and two proteins lacking human homologs. The “consensus” sequence (Lower) was derived from all 116 MIT domains in the SMART database (23). Highly conserved residues (>50% identity) are shown in blue, and residue types conserved in >50% of the sequences are shown in lowercase (h, hydrophobic; +, positively charged; -, negatively charged; u, Ala or Gly; l, Ile, Val, or Leu). Numbering corresponds to VPS4A.
Fig. 2.
VPS4A5–76 MIT domain structure. (A) Ribbon diagram of the VPS4A MIT domain (residues 5–76). (B) View down the three helix bundle of the MIT domain, emphasizing the asymmetry in the disposition of the three helices. (C) “Alanine zipper” connecting VPS4A5–76 MIT helices 1 and 2. The five conserved alanine side chains within this motif are shown explicitly.
Fig. 3.
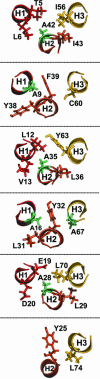
Side chain interactions at the different layers of the VPS4A MIT three-helix bundle. Sequential amino acid layers in the three helix bundle of the VPS4A MIT domain. Note that side chain interactions between helices 2 (H2) and 3 (H3) follow canonical alternating “knobs into holes” interactions between residues at helix positions +1 (A) and +4 (D), whereas side chain packing between helices 1 (H1) and 2 (H2) is different because alternating alanine residues (highlighted in green) project almost directly at the pairing helix and are sandwiched between hydrophobic side chains from the +4 and +5 positions (relative to the preceding alanine) in the pairing helix.
Fig. 4.
Interactions between VPS4A and CHMP1B proteins. (A) GST pull-downs demonstrating that both full-length VPS4A and its isolated MIT domain (VPS4A1–84) bind GST-CHMP1B65–196 (denoted GST-1B65–196, lanes 5 and 7) but not GST alone (lanes 4 and 6). Molecular weight standards (MW, lane 1), pure GST (lane 2), and pure GST-CHMP1B65–196 (lane 3) are shown for reference. (B) Biosensor binding isotherms showing wild-type (WT) and L64A mutant VPS4A1–84 proteins binding GST-CHMP1B and GST-CHMP1B65–196. Filled circles, WT VPS4A1–84 binding full-length GST-CHMP1B captured from E. coli extracts; filled triangles, WT VPS4A1–84 binding GST-CHMP1B65–196 captured from E. coli extracts; filled squares, WT VPS4A1–84 binding affinity purified GST-CHMP1B65–196; open squares, VPS4A1–84 L64A binding affinity purified GST-CHMP1B65–196. Binding to the GST control surface was negligible (not shown). Dissociation constants (μM) for WT VPS4A1–84 binding were: CHMP1B = 20 ± 13 and CHMP1B65–196 = 13 ± 6 μM; and were >500 μM for VPS4A1–84 L64A binding to both constructs (minimum three independent measurements). (C) VPS4A MIT surface renderings showing the locations and identities of conserved residues. Conserved residues are color-coded based on the reduction in CHMP65–196 binding affinity for alanine substitution mutations: dark blue, <3-fold change; light blue, 3- to 30-fold reduction; green, >30-fold reduction. Dissociation constants were: WT, 13 ± 6 μM; I10A, 17 ± 1 μM; K15A, 14 ± 1 μM; E26A, 8 ± 1 μM; E37A, 16 ± 2 μM; K53A, 20 ± 2 μM; R57A, 34 ± 4 μM; K59A, 9 ± 1 μM; L64A, >500 μM; E68A, 55 ± 4 μM; K71A, 18 ± 2 μM (minimum of two independent measurements). (D) Overlays of the first four helices of the TPR domain of FKBP51 (gray) with the MIT domain of VPS4A. The overlay shows the similarity of the structures and the position of the second paired helix (fourth overall) in the FKBP51 TPR domain, which is missing in the VPS4A MIT domain. (E) Side view of the overlay in D, but with the VPS4A MIT domain shown in a space filling model and color coded as in C. The figure emphasizes that residues required for CHMP1B binding (green, light blue) map to the same surface as the binding site for the second paired helix of the FKBP51 TPR domain.
Similar articles
- ESCRT-III recognition by VPS4 ATPases.
Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. Stuchell-Brereton MD, et al. Nature. 2007 Oct 11;449(7163):740-4. doi: 10.1038/nature06172. Nature. 2007. PMID: 17928862 - Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition.
Merrill SA, Hanson PI. Merrill SA, et al. J Biol Chem. 2010 Nov 12;285(46):35428-38. doi: 10.1074/jbc.M110.126318. Epub 2010 Aug 30. J Biol Chem. 2010. PMID: 20805225 Free PMC article. - Binding of Substrates to the Central Pore of the Vps4 ATPase Is Autoinhibited by the Microtubule Interacting and Trafficking (MIT) Domain and Activated by MIT Interacting Motifs (MIMs).
Han H, Monroe N, Votteler J, Shakya B, Sundquist WI, Hill CP. Han H, et al. J Biol Chem. 2015 May 22;290(21):13490-9. doi: 10.1074/jbc.M115.642355. Epub 2015 Apr 1. J Biol Chem. 2015. PMID: 25833946 Free PMC article. - Meiotic Clade AAA ATPases: Protein Polymer Disassembly Machines.
Monroe N, Hill CP. Monroe N, et al. J Mol Biol. 2016 May 8;428(9 Pt B):1897-911. doi: 10.1016/j.jmb.2015.11.004. Epub 2015 Nov 10. J Mol Biol. 2016. PMID: 26555750 Free PMC article. Review. - The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview.
Ahmed I, Akram Z, Iqbal HMN, Munn AL. Ahmed I, et al. Int J Biol Macromol. 2019 Apr 15;127:1-11. doi: 10.1016/j.ijbiomac.2019.01.015. Epub 2019 Jan 4. Int J Biol Macromol. 2019. PMID: 30615963 Review.
Cited by
- Membrane fission reactions of the mammalian ESCRT pathway.
McCullough J, Colf LA, Sundquist WI. McCullough J, et al. Annu Rev Biochem. 2013;82:663-92. doi: 10.1146/annurev-biochem-072909-101058. Epub 2013 Mar 18. Annu Rev Biochem. 2013. PMID: 23527693 Free PMC article. Review. - The ESCRT complexes.
Hurley JH. Hurley JH. Crit Rev Biochem Mol Biol. 2010 Dec;45(6):463-87. doi: 10.3109/10409238.2010.502516. Epub 2010 Jul 23. Crit Rev Biochem Mol Biol. 2010. PMID: 20653365 Free PMC article. Review. - Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH.
Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink HJ, Múnera-Huertas T, Stanton L, Hervás-Aguilar A, Espeso EA, Tilburn J, Arst HN Jr, Peñalva MA. Calcagno-Pizarelli AM, et al. Eukaryot Cell. 2007 Dec;6(12):2365-75. doi: 10.1128/EC.00275-07. Epub 2007 Oct 19. Eukaryot Cell. 2007. PMID: 17951518 Free PMC article. - The molecular mechanism of hepcidin-mediated ferroportin down-regulation.
De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. De Domenico I, et al. Mol Biol Cell. 2007 Jul;18(7):2569-78. doi: 10.1091/mbc.e07-01-0060. Epub 2007 May 2. Mol Biol Cell. 2007. PMID: 17475779 Free PMC article. - Cell biology of the ESCRT machinery.
Hanson PI, Shim S, Merrill SA. Hanson PI, et al. Curr Opin Cell Biol. 2009 Aug;21(4):568-74. doi: 10.1016/j.ceb.2009.06.002. Epub 2009 Jun 26. Curr Opin Cell Biol. 2009. PMID: 19560911 Free PMC article. Review.
References
- Murk, J. L., Stoorvogel, W., Kleijmeer, M. J. & Geuze, H. J. (2002) Semin. Cell Dev. Biol. 13, 303–311. - PubMed
- Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893–905. - PubMed
- Gruenberg, J. & Stenmark, H. (2004) Nat. Rev. Mol. Cell Biol. 5, 317–323. - PubMed
- de Gassart, A., Geminard, C., Hoekstra, D. & Vidal, M. (2004) Traffic 5, 896–903. - PubMed
- Morita, E. & Sundquist, W. I. (2004) Annu. Rev. Cell Dev. Biol. 20, 395–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases