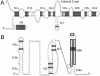Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains - PubMed (original) (raw)
Comparative Study
. 2005 Oct 4;102(40):14249-54.
doi: 10.1073/pnas.0503838102. Epub 2005 Sep 23.
Affiliations
- PMID: 16183745
- PMCID: PMC1242290
- DOI: 10.1073/pnas.0503838102
Comparative Study
Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains
Christophe Pichon et al. Proc Natl Acad Sci U S A. 2005.
Erratum in
- Proc Natl Acad Sci U S A. 2005 Nov 15;102(46):16905
Abstract
Small RNA (sRNA) genes are expressed in all organisms, primarily as regulators of translation and message stability. We have developed comparative genomic approaches to identify sRNAs that are expressed by Staphylococcus aureus, the most common cause of hospital-acquired infections. This study represents an in-depth analysis of the RNome of a Gram-positive bacterium. A set of sRNAs candidates were identified in silico within intergenic regions, and their expression levels were monitored by using microarrays and confirmed by Northern blot hybridizations. Two sRNAs were also detected directly from purification and RNA sequence determination. In total, at least 12 sRNAs are expressed from the S. aureus genome, five from the core genome and seven from pathogenicity islands that confer virulence and antibiotic resistance. Three sRNAs are present in multiple (two to five) copies. For the sRNAs that are conserved throughout the bacterial phylogeny, their secondary structures were inferred by phylogenetic comparative methods. In vitro binding assays indicate that one sRNA encoded within a pathogenicity island is a trans-encoded antisense RNA regulating the expression of target genes at the posttranscriptional level. Some of these RNAs show large variations of expression among pathogenic strains, suggesting that they are involved in the regulation of staphylococcal virulence.
Figures
Fig. 1.
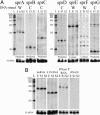
Experimental identification of highly expressed sRNAs in S. aureus. (A) Total RNA extracted at exponential (E) or early stationary (S1) phase analyzed by 8% PAGE stained by ethidium bromide (L, RNA ladder). (B) Three unknown RNAs or RNA fragments (1-3) purified and sequenced with RNases T1 (GL) and U2 (AL). C, controls; L, hydrolysis RNA ladders; 12% PAGE for 1 and 2, 8% PAGE for 3. The RNA sequence is indicated, Y are the pyrimidines. (C) Total RNA (15 μg per lane) analyzed on a Northern blot by using labeled DNA probes complementary to the previously uncharacterized sRNAs, with a 100- to 1,000-nt-long RNA ladder. As a control, the blot was probed for 5S rRNA.
Fig. 2.
Experimental verification of the predicted sRNAs in S. aureus. Northern hybridization was performed by using strand-specific probes. (A) The labeled probes target either an sRNA candidate expressed from the Watson (W) or the Crick (C) strands. Total RNA were extracted from N315 cells grown under exponential (E), early (S1), and late (S2) stationary phases. The signal intensity gives an indication of the relative abundance between sRNAs (exposure times are similar for each image). (B) tmRNA, 4.5S RNA, and RNase P RNA are expressed in S. aureus during cell growth. Strain N315 does not express RNAIII. 5′ end-labeled 100- to 1,000-nt RNA molecular weight markers allow a direct estimation of RNA transcript length. As loading controls, the blots were probed for the 5S rRNA.
Fig. 3.
Copy number and genomic location of the sRNA genes in S. aureus. (A) The copy number of the sRNA genes within the genomic sequences of S. aureus N315, MRSA252, and COL; in brackets are the locations of the sRNA genes. tmRNA, 4.5S RNA, and RNase P RNA are in single copies per genome. (B) Schematic view of the genome of S. aureus N315 with emphasis (rectangles) to three pathogenicity islands (SaPI 1-SaPI 3), a staphylococcal cassette chromosome (SCC), a bacteriophage (φN315), and the location of the detected sRNA genes. The location of the multiple copies of sprA and sprF-G identified in N315 is indicated. Nucleotide position, in Mb, is indicated. (C) Identity of the genes flanking the sRNA genes. The genes that are involved in resistance or virulence functions are in bold. The location on S. aureus N315 chromosome is indicated. HP, hypothetical protein.
Fig. 4.
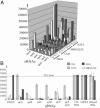
Comparative expression profiles of S. aureus sRNAs across microarrays. (A) Three-dimensional diagram of the normalized intensity of expression of sRNAs sprA-sprFG among four strains of S. aureus at exponential (E), early (S1), and late (S2) stationary phases (B) Comparative normalized expression profiles of 11 sRNAs in four S. aureus strains at growth phase S1. The expression levels below the lower horizontal line are considered as insignificant. The error bars are indicated.
Fig. 5.
Expression profiles of the sRNAs among four S. aureus pathogenic strains. Northern hybridization was performed by using strand-specific probes. (A) RNAIII is expressed only in MRSA252 (weakly) and 502A (strongly), and 6S RNA is expressed in all four strains. (B and C) Expression profiles of seven sRNAs located in S. aureus pathogenicity islands. RNA markers allow a direct estimation of RNA transcript length. As a loading control, the blots were probed for 5S rRNA.
Fig. 6.
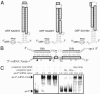
Secondary structures of S. aureus 6S RNA (A) and sprA (B) based on comparative sequence analysis (
supporting information
). Phylogenetic support for H1ab and H4a in sprA is weak. The bars are the GC and AU pairs, and the dotted lines are the GU pairs. In sprA, the boxed nucleotides are predicted to interact with target mRNAs through antisense pairings.
Fig. 7.
SprA has antisense functions and targets selected mRNA 3′ UTRs. (A) Three mRNA 3′ UTRs have sequence complementarities with sequences at the 3′ end of sprA (H4b). The ORFs are numbered according to ref. . The termination codons of the mRNAs are indicated; (N)x are the nucleotide linking the stop codons to the predicted hairpins containing the nucleotide (boxed) that can pair with the nucleotide from sprA (in H4b). (B) Proposed GC and AU pairs between the nucleotide from sprA with the nucleotide from the 3′ UTRs of the mRNA targets. The two triangles indicate the position of nucleotides that are missing in ORF SA1944, but the other pairings are maintained. Sequence variation is extensive in the unpaired regions of the mRNA sequences. (C) Specific interaction between SprA and an mRNA fragment derived from ORF SA2216 (a predicted ABC transporter) in vitro. Complex formation between the two RNAs is detected by native gel retardation assay of labeled sprA with increasing amounts of unlabeled mRNA. The addition of either unlabeled sprA or bulk tRNA from yeast demonstrates specificity.
Similar articles
- Identification of bacterial small non-coding RNAs: experimental approaches.
Altuvia S. Altuvia S. Curr Opin Microbiol. 2007 Jun;10(3):257-61. doi: 10.1016/j.mib.2007.05.003. Epub 2007 Jun 5. Curr Opin Microbiol. 2007. PMID: 17553733 Review. - A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule.
Chabelskaya S, Gaillot O, Felden B. Chabelskaya S, et al. PLoS Pathog. 2010 Jun 3;6(6):e1000927. doi: 10.1371/journal.ppat.1000927. PLoS Pathog. 2010. PMID: 20532214 Free PMC article. - Staphylococcus aureus multiresistance plasmid pSK41: analysis of the replication region, initiator protein binding and antisense RNA regulation.
Kwong SM, Skurray RA, Firth N. Kwong SM, et al. Mol Microbiol. 2004 Jan;51(2):497-509. doi: 10.1046/j.1365-2958.2003.03843.x. Mol Microbiol. 2004. PMID: 14756789 - Identification of essential genes in Staphylococcus aureus by construction and screening of conditional mutant library.
Yin D, Ji Y. Yin D, et al. Methods Mol Biol. 2008;416:297-305. doi: 10.1007/978-1-59745-321-9_19. Methods Mol Biol. 2008. PMID: 18392975 - How to find small non-coding RNAs in bacteria.
Vogel J, Sharma CM. Vogel J, et al. Biol Chem. 2005 Dec;386(12):1219-38. doi: 10.1515/BC.2005.140. Biol Chem. 2005. PMID: 16336117 Review.
Cited by
- Developing New Tools to Fight Human Pathogens: A Journey through the Advances in RNA Technologies.
Costa VG, Costa SM, Saramago M, Cunha MV, Arraiano CM, Viegas SC, Matos RG. Costa VG, et al. Microorganisms. 2022 Nov 21;10(11):2303. doi: 10.3390/microorganisms10112303. Microorganisms. 2022. PMID: 36422373 Free PMC article. Review. - No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus.
Bohn C, Rigoulay C, Bouloc P. Bohn C, et al. BMC Microbiol. 2007 Feb 9;7:10. doi: 10.1186/1471-2180-7-10. BMC Microbiol. 2007. PMID: 17291347 Free PMC article. - Insights Into the Impact of Small RNA SprC on the Metabolism and Virulence of Staphylococcus aureus.
Zhou J, Zhao H, Yang H, He C, Shu W, Cui Z, Liu Q. Zhou J, et al. Front Cell Infect Microbiol. 2022 Feb 23;12:746746. doi: 10.3389/fcimb.2022.746746. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35281456 Free PMC article. - The staphylococcus-specific gene rsr represses agr and virulence in Staphylococcus aureus.
Tamber S, Reyes D, Donegan NP, Schwartzman JD, Cheung AL, Memmi G. Tamber S, et al. Infect Immun. 2010 Oct;78(10):4384-91. doi: 10.1128/IAI.00401-10. Epub 2010 Aug 9. Infect Immun. 2010. PMID: 20696829 Free PMC article. - Identification of small Hfq-binding RNAs in Listeria monocytogenes.
Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. Christiansen JK, et al. RNA. 2006 Jul;12(7):1383-96. doi: 10.1261/rna.49706. Epub 2006 May 8. RNA. 2006. PMID: 16682563 Free PMC article.
References
- Storz, G., Opdyke, J. A. & Zhang, A. (2004) Curr. Opin. Microbiol. 7, 140-144. - PubMed
- Johansson, J. & Cossart, P. (2003) Trends Microbiol. 11, 280-285. - PubMed
- Kwong, S. M., Skurray, R. A. & Firth, N. (2004) Mol. Microbiol. 51, 497-509. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases