Coronaviruses as vectors: stability of foreign gene expression - PubMed (original) (raw)
Comparative Study
Coronaviruses as vectors: stability of foreign gene expression
Cornelis A M de Haan et al. J Virol. 2005 Oct.
Abstract
Coronaviruses are enveloped, positive-stranded RNA viruses considered to be promising vectors for vaccine development, as (i) genes can be deleted, resulting in attenuated viruses; (ii) their tropism can be modified by manipulation of their spike protein; and (iii) heterologous genes can be expressed by simply inserting them with appropriate coronaviral transcription signals into the genome. For any live vector, genetic stability is an essential requirement. However, little is known about the genetic stability of recombinant coronaviruses expressing foreign genes. In this study, the Renilla and the firefly luciferase genes were systematically analyzed for their stability after insertion at various genomic positions in the group 1 coronavirus feline infectious peritonitis virus and in the group 2 coronavirus mouse hepatitis virus. It appeared that the two genes exhibit intrinsic differences, the Renilla gene consistently being maintained more stably than the firefly gene. This difference was not caused by genome size restrictions, by different effects of the encoded proteins, or by different consequences of the synthesis of the additional subgenomic mRNAs. The loss of expression of the firefly luciferase was found to result from various, often large deletions of the gene, probably due to RNA recombination. The extent of this process appeared to depend strongly on the coronaviral genomic background, the luciferase gene being much more stable in the feline than in the mouse coronavirus genome. It also depended significantly on the particular genomic location at which the gene was inserted. The data indicate that foreign sequences are more stably maintained when replacing nonessential coronaviral genes.
Figures
FIG. 1.
Genomic organization of the recombinant viruses. The genome structures of the recombinant wild-type MHV-A59 (MHV-WT) and FIPV (FIPV-WT) viruses and of the recombinant viruses containing either an RL or an FL expression cassette or both are depicted. The asterisks indicate either the position of a premature stop codon (MHV-EFLMSTOP) or of a mutated TRS (MHV-EFLMTRS). Numbers designate the genes encoding nonstructural proteins, while genes encoding the hemagglutinin-esterase (HE) protein, spike (S) protein, envelope (E) protein, membrane (M) protein, or nucleocapsid (N) protein are indicated by their abbreviations. The 5′- and 3′-untranslated regions (UTR) are also designated.
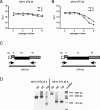
FIG. 2.
Stability of expression of RL versus FL within an otherwise identical background. Recombinant viruses MHV-ERLM (A) and MHV-EFLM (B) were passaged eight times on LR7 cells as described in Materials and Methods. Subsequently, in a parallel expression experiment, LR7 cells were infected with the recombinant viruses from the different passages at an MOI of 8. At 8.5 h postinfection, the cells were lysed and the intracellular luciferase expression was determined by using a luminometer (values are expressed in relative light units [RLU]). Independent recombinants are indicated with the letters A and B. (C) RT-PCR was used to amplify regions of genomic RNA. The RT step was performed with primer 746 (5′-CGTCTAGATTAGGTTCTCAACAATGCGG-3′), which is complementary to the 3′ end of the M gene, while the PCR was performed with primers 509 and 1091 (5′-GTTACAAACCTGAATCTCATCTTAATTCTGGTCG-3′), which are complementary to sequences in the 3′ end of the M gene and in the 5a gene, respectively. The same strategy was employed for the RL- and FL-expressing viruses. The sizes of the expected full-length PCR products are indicated. (D) The RT-PCR analysis was performed for both MHV-ERLM (recombinant A) and MHV-EFLM (recombinant A) from passages 2 (P2), 5 (P5), and 8 (P8). PCR products were analyzed by electrophoresis in 1% agarose gels stained with ethidium bromide. Sizes of the relevant DNA fragments of the marker are indicated.
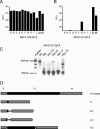
FIG. 3.
Analysis of cloned MHV-ERLM and MHV-EFLM after serial passaging. Passage 8 viruses from MHV-ERLM (A) and MHV-EFLM (B) were plaque purified once, after which stocks were grown on LR7 cells (passage 8.1, A to J). Intracellular luciferase expressed in LR7 cells after infection with these plaque-purified viruses was determined. Passage 2 (p2) and passage 8 (p8) viruses were used as references. (C) RT-PCR was used to amplify regions of genomic RNA as described in the legend to Fig. 2. The analysis was performed for MHV-EFLM (recombinant A) from passage 2 (P2), from passage 8 (P8), and from the plaque-purified viruses (P8.1, E to G). Sizes of the relevant DNA fragments of the marker are indicated. (D) The PCR products were cloned and sequenced. A schematic representation of the sequencing results is shown. Genes E and M are indicated by gray boxes, while the inserted expression cassette is shown in black. The open reading frame that will be translated theoretically from the inserted sequences (or what is left of it) is indicated with a black box. The plaques from which these sequences were derived are indicated on the right.
FIG. 4.
Sequence analysis of the viruses plaque purified from MHV-EFLM passage 8. The detailed sequencing results from the MHV-EFLM viruses derived from plaques E, D, F, and G are shown. The 5′ end of the E gene and the 3′ end of the M gene are indicated as well as the encoded amino acid sequence. The sequence of the inserted expression cassette is shown in boldface (nt 1 to 1691). The TRSs are indicated. Viruses derived from plaque D only contain the first 56 nt from the expression cassette fused in frame to the 3′ end of the M gene. Viruses derived from plaques E and F had lost nt 43 to 1576 and 1597 to 1650 from the expression cassette. Two sequences were obtained from plaque G (G1 and G2). The G.1 sequence shows various deletions as well as rearrangements in the expression cassette. As a result, the E gene has been extended with a few residues. The G.2 sequence shows one large deletion of nt 944 through 1624.
FIG. 5.
Stability of foreign gene expression of MHV-minFL. (A) LR7 cells were infected at an MOI of 8. At 8.5 h postinfection, the viral infectivity in the culture medium was determined by a quantal assay on LR7 cells, and TCID50 values were calculated. (B) In the same experiment the intracellular expression of the FL gene (in RLU) was determined by using a luminometer. Standard deviations are indicated. (C) The stability of foreign gene expression upon serial passaging was determined as described in the legend to Fig. 2 and compared to that of MHV-EFLM (D). The analysis of individual clones of MHV-minFL (passage 8) was performed as described in the legend to Fig. 3.
FIG. 6.
Stability of foreign gene expression of other recombinant viruses. (A) The sequences of the 3′ end of the FL expression cassettes of MHV-EFLM, MHV-EFLMSTOP, and MHV-EFLMTRS are shown. The asterisk represents a stop codon. The TRSs are indicated. The single nucleotide deleted in MHV-EFLMSTOP is shown in boldface and is indicated with an arrow in the sequence of MHV-EFLM. The nucleotides mutated in the TRS in MHV-EFLMTRS are shown in boldface and are not underlined. MHV-EFLMSTOP (B), MHV-EFLMTRS (C), and MHV-2aFLS (D) were passaged eight times as described in Materials and Methods. The viruses from passage 8 were analyzed as described in the legend to Fig. 3. As a reference, the intracellular FL expression of MHV-EFLM (passage 2) is also shown (FL). (E) MHV-FLRL and MHV-RLFL were passaged eight times as described, and viruses from passages 2, 5, and 8 were analyzed. LR7 cells were infected at an MOI of 8. At 8.5 h postinfection, the intracellular expression of RL and FL was determined, and the FL/RL ratio was calculated. Standard deviations are indicated.
FIG. 7.
Stability of FIPV foreign gene expression. (A and B) Single-step growth kinetics of FIPV recombinants. FCWF cells were infected with each recombinant FIPV at an MOI of 8. Virus infectivity in the culture medium was determined at different times postinfection by a quantal assay on LR7 cells, and TCID50 values were calculated. Independently generated recombinants are indicated with the letters A and B. (C and D) In the same experiment, the intracellular expression of RL or FL (in relative light units [RLU]) was determined by using a luminometer. (E and F) The stability of foreign gene expression upon serial passaging was determined as described in the legend to Fig. 2, except that FCWF cells were used. (G and H) The analysis of individual clones of passage 8 viruses was performed as described in the legend to Fig. 3, except that FCWF cells were used. WT, wild type.
Similar articles
- Coronaviruses as vectors: position dependence of foreign gene expression.
de Haan CA, van Genne L, Stoop JN, Volders H, Rottier PJ. de Haan CA, et al. J Virol. 2003 Nov;77(21):11312-23. doi: 10.1128/jvi.77.21.11312-11323.2003. J Virol. 2003. PMID: 14557617 Free PMC article. - Switching species tropism: an effective way to manipulate the feline coronavirus genome.
Haijema BJ, Volders H, Rottier PJ. Haijema BJ, et al. J Virol. 2003 Apr;77(8):4528-38. doi: 10.1128/jvi.77.8.4528-4538.2003. J Virol. 2003. PMID: 12663759 Free PMC article. - Manipulation of the coronavirus genome using targeted RNA recombination with interspecies chimeric coronaviruses.
de Haan CA, Haijema BJ, Masters PS, Rottier PJ. de Haan CA, et al. Methods Mol Biol. 2008;454:229-36. doi: 10.1007/978-1-59745-181-9_17. Methods Mol Biol. 2008. PMID: 19057874 Free PMC article. - Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis.
Tekes G, Thiel HJ. Tekes G, et al. Adv Virus Res. 2016;96:193-218. doi: 10.1016/bs.aivir.2016.08.002. Epub 2016 Aug 31. Adv Virus Res. 2016. PMID: 27712624 Free PMC article. Review. - Coronavirus reverse genetics and development of vectors for gene expression.
Enjuanes L, Sola I, Alonso S, Escors D, Zúñiga S. Enjuanes L, et al. Curr Top Microbiol Immunol. 2005;287:161-97. doi: 10.1007/3-540-26765-4_6. Curr Top Microbiol Immunol. 2005. PMID: 15609512 Free PMC article. Review.
Cited by
- Development of HiBiT-Tagged Recombinant Infectious Bronchitis Coronavirus for Efficient in vitro and in vivo Viral Quantification.
Liang XY, Zhu QC, Liang JQ, Liu SY, Liu DX, Fung TS. Liang XY, et al. Front Microbiol. 2020 Aug 28;11:2100. doi: 10.3389/fmicb.2020.02100. eCollection 2020. Front Microbiol. 2020. PMID: 32983065 Free PMC article. - Modified H5 promoter improves stability of insert genes while maintaining immunogenicity during extended passage of genetically engineered MVA vaccines.
Wang Z, Martinez J, Zhou W, La Rosa C, Srivastava T, Dasgupta A, Rawal R, Li Z, Britt WJ, Diamond D. Wang Z, et al. Vaccine. 2010 Feb 10;28(6):1547-57. doi: 10.1016/j.vaccine.2009.11.056. Epub 2009 Dec 5. Vaccine. 2010. PMID: 19969118 Free PMC article. - Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.
Drexler JF, Corman VM, Drosten C. Drexler JF, et al. Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review. - ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells.
Burkard C, Verheije MH, Haagmans BL, van Kuppeveld FJ, Rottier PJ, Bosch BJ, de Haan CA. Burkard C, et al. J Virol. 2015 Apr;89(8):4434-48. doi: 10.1128/JVI.03274-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653449 Free PMC article. - Transmissible gastroenteritis virus (TGEV)-based vectors with engineered murine tropism express the rotavirus VP7 protein and immunize mice against rotavirus.
Ribes JM, Ortego J, Ceriani J, Montava R, Enjuanes L, Buesa J. Ribes JM, et al. Virology. 2011 Feb 5;410(1):107-18. doi: 10.1016/j.virol.2010.10.036. Epub 2010 Nov 21. Virology. 2011. PMID: 21094967 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources