Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates - PubMed (original) (raw)
Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates
Cerstin Franz et al. EMBO J. 2005.
Abstract
Nuclear envelope (NE) formation during cell division in multicellular organisms is a central yet poorly understood biological process. We report that the conserved nucleoporin Nup155 has an essential function in NE formation in Caenorhabditis elegans embryos and in Xenopus laevis egg extracts. In vivo depletion of Nup155 led to failure of nuclear lamina formation and defects in chromosome segregation at anaphase. Nup155 depletion inhibited accumulation of nucleoporins at the nuclear periphery, including those recruited to chromatin early in NE formation. Electron microscopy analysis revealed that Nup155 is also required for the formation of a continuous nuclear membrane in vivo and in vitro. Time-course experiments indicated that Nup155 is recruited to chromatin at the time of NE sealing, suggesting that nuclear pore complex assembly has to progress to a relatively late stage before NE membrane assembly occurs.
Figures
Figure 1
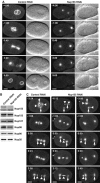
Depletion of Nup155 from C. elegans embryos affects nuclear appearance. (A) Embryos expressing GFP-β-tubulin and YFP-lamin from control (left panels) or Nup155-depleted worms (right panels) were observed by time-lapse microscopy. In this and other figures, images are shown with indication of time (minutes:seconds) relative to anaphase onset and with embryos anterior to the left. In the Nup155 RNAi embryo, neither pronuclei nor nuclei were visible. See Supplementary video 4. (B) Western blot analysis of embryonic lysates from mock-depleted (left lane) or Nup155-depleted worms (right lane) with antisera against various nucleoporins. The asterisk marks a protein that crossreacts with the Nup155 antibody, and demonstrates equal gel loading. (C) Control (left) or Nup155-depleted (middle and right) embryos expressing GFP-β-tubulin and GFP-histone H2B were observed by time-lapse microscopy. In selected panels, centrosomes are indicated with arrows whereas triangles point to chromatin. See Supplementary video 5. Scale bars, 10 μm.
Figure 2
C. elegans Nup155 is required for NPC assembly. (A) Control (upper panels) or Nup155-depleted (lower panels) embryos were fixed and analysed using anti-Nup153 antibodies (green in overlay) or mAb414 (red in overlay). Chromatin was visualized with DAPI (blue in overlay). (B) Control (upper panels) or Nup155-depleted (lower panels) embryos were analysed with anti-Nup96 (left two columns, green in overlay) or anti-Nup35 (right two columns, green in overlay) antibodies together with mAb414 and DAPI (red and blue in overlay, respectively). (C) Still images from time-lapse recordings of YFP-Nup107-expressing control embryos (left panels) or Nup155-depleted embryos (right panels). The inset in left panel shows a magnification of YFP-Nup107 localization to the metaphase plate (−0:20). See Supplementary video 6. Scale bars, 10 μm.
Figure 3
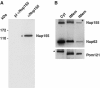
Biochemical characterization of Nup155 in Xenopus egg extract. (A) The cytosolic fraction of Xenopus egg extracts was analysed by Western blotting with preimmune (pi αNup155) or anti-Nup155 sera (αNup155). Molecular weight markers are indicated on the left. (B) Equal amounts of protein of the cytosol (Cyt), total membrane (tMem) and floated membrane (fMem) fractions of Xenopus egg extract were probed for the antigens Nup155, Nup62 and Pom121 on a Western blot. The asterisk in the cytosol lane indicates a crossreactivity of the Pom121 antibody.
Figure 4
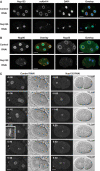
Immunodepletion of Nup155 inhibits NPC formation in vitro. (A) Nup155 was immunodepleted from cytosol and nuclei were assembled in vitro. Nucleoporin localization was analysed by immunofluorescence in mock-depleted (Mock) and Nup155-depleted reactions (ΔNup155), or after readdition of recombinant Nup155 (ΔNup155+Nup155). Nup155, mAb414 and overlay (Nup155 (green), mAb414 (red) and DAPI in blue) are costainings of the same nuclei. Scale bars, 5 μm. (B) Amounts of Nup155 in mock-depleted (Mock) and Nup155-depleted (ΔNup155) cytosol was compared by Western blotting. Equal volumes were loaded while varying amounts of untreated cytosol (Cyt) were loaded for comparison. A 0.6 μl portion of cytosol (25 μg total protein) corresponds to 50% of the sample volume loaded of mock- and Nup155-depleted cytosol. Tubulin serves as a loading control. (C, D) As in panel A, but co-immunodetection of Nup107 (C) or Pom121 (D) with mAb414 antigens.
Figure 5
Membrane defects upon Nup155 depletion. (A) Control (left panels) or Nup155-depleted (right panels) embryos expressing GFP-MAN1 were observed by time-lapse microscopy. Centrosomes are indicated with arrows at anaphase (0:40) while triangles point to GFP-MAN1 association with chromatin during telophase (2:40). See Supplementary video 7. Scale bars, 10 μm. (B) Nuclei assembled in vitro in mock- or Nup155-depleted extracts, or in depleted extracts after addition of recombinant Nup155, were labelled with the membrane dye DiIC18 before fixation in 2% paraformaldehyde and 0.5% glutaraldehyde and analysed by confocal microscopy. Left and right image sets were derived from independent experiments. The two left panels are individually adjusted in signal intensity to illustrate the nature of the membranes best possible. Equal exposure times were chosen for right images. (C) Quantitation of closed NEs from panel B. In three independent experiments, 100 nuclei from each reaction were examined. The average of the three independent experiments is shown; error bars represent the total variation over the experiments.
Figure 6
TEM analysis of NE defects in vivo and in vitro. (A, B) Control (A) or Nup155-depleted (B) C. elegans embryos were fixed by high-pressure freezing and processed for TEM. White boxes in upper images indicate the area shown at higher magnification below. In the higher magnifications, chromatin is localized to the lower right half of the micrographs. In panel A, empty triangles point to NPCs, which were not visible in Nup155 RNAi samples. In panel B, arrows point to areas where the chromatin surface is not covered by membranes while filled triangles indicate chromatin surface that is covered by membranes without NPCs. An asterisk indicates a membrane vesicle or tubule present inside the chromatin space. Scale bars, 0.5 μm (upper images) and 0.1 μm (lower images). (C–E) _In vitro_-assembled nuclei as in Figure 4A were processed for TEM. (C) Mock depleted. Scale bar, 0.2 μm. (D) Nup155 depleted. Scale bars, 2 μm (upper image) and 0.5 μm (middle and lower images). (E) Nup155 depleted followed by addition of recombinant Nup155. Scale bars, 1 μm (upper image) and 0.2 μm (middle and lower images).
Figure 7
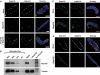
Dynamics of Nup155. (A) Still images from time-lapse microscopy recording of C. elegans embryo expressing GFP-Nup155. Images show progression from pronuclear meeting in the one-cell embryo to the two-cell stage. The inset shows a magnification of initial GFP-Nup155 recruitment to the two reforming nuclei during anaphase (1:20). See Supplementary video 8. Scale bar, 10 μm. (B) NE assembly reactions were fixed in 4% paraformaldehyde at the time points indicated. The three columns on the left, which show immunodetection of Nup155 (green), mAb414 (red) and DAPI (blue), are identical samples and merged in the overlaid images. Parallel reactions for Nup107 immunodetection or that were stained with the membrane dye DiIC18 before fixation are on the right. Scale bars, 5 μm.
Similar articles
- Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly.
Ródenas E, Klerkx EP, Ayuso C, Audhya A, Askjaer P. Ródenas E, et al. Dev Biol. 2009 Mar 15;327(2):399-409. doi: 10.1016/j.ydbio.2008.12.024. Epub 2008 Dec 30. Dev Biol. 2009. PMID: 19146848 - Channel Nucleoporins Recruit PLK-1 to Nuclear Pore Complexes to Direct Nuclear Envelope Breakdown in C. elegans.
Martino L, Morchoisne-Bolhy S, Cheerambathur DK, Van Hove L, Dumont J, Joly N, Desai A, Doye V, Pintard L. Martino L, et al. Dev Cell. 2017 Oct 23;43(2):157-171.e7. doi: 10.1016/j.devcel.2017.09.019. Dev Cell. 2017. PMID: 29065307 Free PMC article. - Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo.
Galy V, Mattaj IW, Askjaer P. Galy V, et al. Mol Biol Cell. 2003 Dec;14(12):5104-15. doi: 10.1091/mbc.e03-04-0237. Epub 2003 Aug 22. Mol Biol Cell. 2003. PMID: 12937276 Free PMC article. - What can Caenorhabditis elegans tell us about the nuclear envelope?
Gorjánácz M, Jaedicke A, Mattaj IW. Gorjánácz M, et al. FEBS Lett. 2007 Jun 19;581(15):2794-801. doi: 10.1016/j.febslet.2007.03.052. Epub 2007 Mar 30. FEBS Lett. 2007. PMID: 17418822 Review. - Chromatin organization in relation to the nuclear periphery.
Kalverda B, Röling MD, Fornerod M. Kalverda B, et al. FEBS Lett. 2008 Jun 18;582(14):2017-22. doi: 10.1016/j.febslet.2008.04.015. Epub 2008 Apr 22. FEBS Lett. 2008. PMID: 18435921 Review.
Cited by
- Ndc1 drives nuclear pore complex assembly independent of membrane biogenesis to promote nuclear formation and growth.
Mauro MS, Celma G, Zimyanin V, Magaj MM, Gibson KH, Redemann S, Bahmanyar S. Mauro MS, et al. Elife. 2022 Jul 19;11:e75513. doi: 10.7554/eLife.75513. Elife. 2022. PMID: 35852146 Free PMC article. - Pan-cancer analysis of NUP155 and validation of its role in breast cancer cell proliferation, migration, and apoptosis.
Wang ZQ, Wu ZX, Wang ZP, Bao JX, Wu HD, Xu DY, Li HF, Xu YY, Wu RX, Dai XX. Wang ZQ, et al. BMC Cancer. 2024 Mar 19;24(1):353. doi: 10.1186/s12885-024-12039-6. BMC Cancer. 2024. PMID: 38504158 Free PMC article. - Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin.
Schwartz M, Travesa A, Martell SW, Forbes DJ. Schwartz M, et al. Nucleus. 2015;6(1):40-54. doi: 10.1080/19491034.2015.1004260. Nucleus. 2015. PMID: 25602437 Free PMC article. - Structure, dynamics and function of nuclear pore complexes.
D'Angelo MA, Hetzer MW. D'Angelo MA, et al. Trends Cell Biol. 2008 Oct;18(10):456-66. doi: 10.1016/j.tcb.2008.07.009. Epub 2008 Sep 9. Trends Cell Biol. 2008. PMID: 18786826 Free PMC article. Review. - Transmembrane protein-free membranes fuse into xenopus nuclear envelope and promote assembly of functional pores.
Rafikova ER, Melikov K, Ramos C, Dye L, Chernomordik LV. Rafikova ER, et al. J Biol Chem. 2009 Oct 23;284(43):29847-59. doi: 10.1074/jbc.M109.044453. Epub 2009 Aug 20. J Biol Chem. 2009. PMID: 19696024 Free PMC article.
References
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW (2005) The integral membrane nucleoporin Pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17: 83–92 - PubMed
- Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B (1999) Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci 112: 2253–2264 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous