Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex - PubMed (original) (raw)
Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex
David R Peaper et al. EMBO J. 2005.
Abstract
We previously showed that the major histocompatibility complex (MHC) class I chaperone tapasin can be detected as a mixed disulfide with the thiol-oxidoreductase ERp57. Here we show that tapasin is a unique and preferred substrate, a substantial majority of which is disulfide-linked to ERp57 within the cell. Tapasin upregulation by interferon-gamma induces sequestration of the vast majority of ERp57 into the MHC class I peptide-loading complex. The rate of tapasin-ERp57 conjugate formation is unaffected by the absence of beta2-microglubulin (beta2m), and is independent of calnexin or calreticulin interactions with monoglucosylated N-linked glycans. The heterodimer forms spontaneously in vitro upon mixing recombinant ERp57 and tapasin. Noncovalent interactions between the native proteins inhibit the reductase activity of the thioredoxin CXXC motif within the N-terminal a domain of ERp57 to maintain its interaction with tapasin. Disruption of these interactions by denaturation allows reduction to proceed. Thus, tapasin association specifically inhibits the escape pathway required for disulfide-bond isomerization within conventional protein substrates, suggesting a specific structural role for ERp57 within the MHC class I peptide-loading complex.
Figures
Figure 1
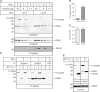
IFN-γ treatment drives tapasin association and decreases the pool of free ERp57. (A) IFN-γ reduces the pool of free ERp57. HeLa-M cells were treated with IFN-γ for 2 days prior to harvesting and MMTS treatment. Three-fold serial dilutions of cell extracts were resolved by SDS–PAGE under reducing or nonreducing conditions and blotted with rabbit anti-ERp57 raised against a C-terminal peptide. After probing for ERp57, membranes were stripped, cut, and re-probed for tapasin with anti-gp48N or for ERp72 as a loading control. The tapasin/ERp57 conjugate and free ERp57 are indicated. (B) IFN-γ reduces the pool of free ERp57. Bands in (A) corresponding to conjugated and reduced ERp57 were quantitated, and the percent of conjugated ERp57 was calculated as Percent Conjugated ERp57=(Conjugated ERp57/Reduced ERp57)*100. Data shown are the average±s.e.m. of three dilutions from two separate experiments. (C) IFN-γ increases conjugate levels to the same extent as tapasin expression. Bands in (A) corresponding to tapasin and conjugated ERp57 with and without IFN-γ treatment were quantitated, and the fold increase in expression was calculated. Data shown are the average ± s.e.m. of three dilutions from two separate experiments. (D) IFN-γ induction of tapasin reduces the pool of free ERp57. Lysates from (A) were precleared and precipitated with PaSta1, MaP.ERp57, or M.PDI and protein G Sepharose. Samples were resolved under reducing or nonreducing conditions as indicated and blotted with rabbit anti-peptide R.ERp57-C. The upper molecular weight species contains both tapasin and ERp57, and ERp57 exists in at least two oxidation states. The tapasin/ERp57 conjugate and free ERp57 are indicated. (E) All detectable tapasin is conjugated to ERp57 under steady-state conditions. HeLa-M cells were treated with IFN-γ for 2 days prior to harvesting and MMTS treatment. Triton X-100 extracts were resolved by SDS–PAGE under reducing or nonreducing conditions and probed for tapasin and ERp57 as in (A). Membranes were stripped and reblotted for ERp72 as a loading control. Samples were visualized with fluorimetry. The tapasin/ERp57 conjugate, free ERp57, and reduced tapasin are indicated.
Figure 2
Conjugate formation is independent of β2m. All TAP-associated and cellular tapasin is conjugated to ERp57 in the presence or absence of β2m. Post-nuclear supernatants from IFN-γ-treated FO-1 or FO-1.β2m cells solubilized in the indicated detergents were precipitated with 148.3, MaP.ERp57, PaSta-1, or M.PDI and protein G Sepharose. SDS–PAGE was performed under reducing or nonreducing conditions and blots were probed for tapasin with R.gp48C. The tapasin/ERp57 conjugate and reduced tapasin are indicated.
Figure 3
β2m expression does not affect the rate of conjugate formation. (A, B) IFN-γ-treated FO-1 or FO-1.β2m cells pulsed with [35S]methionine and cysteine for 5 min were chased for the indicated periods prior to solubilization in digitonin. Post-nuclear supernatants were precipitated with the anti-TAP mAb 148.3 or mouse IgG coupled to agarose beads (C=Ctrl). Associated proteins were stripped in 1% SDS, and tapasin and its conjugate with ERp57 were reprecipitated with R.gp48C and protein A Sepharose. The tapasin/ERp57 conjugate, free ERp57, and free tapasin are indicated. (C) Data from panels A and B are presented as a percentage of the maximum ERp57–tapasin conjugate signal throughout the chase period. (D) Newly synthesized tapasin represents most of the radioactivity incorporated into the conjugate. Samples were prepared as in (A) and (B), but gels were run under reducing conditions. Reduced ERp57 and tapasin are indicated.
Figure 4
Tapasin completes oxidative folding after TAP association. IFN-γ-stimulated FO-1.β2m cells pulsed with [35S]methionine and cysteine for 5 min were chased for the indicated times prior to solubilization in digitonin. Post-nuclear supernatants were precipitated with an anti-TAP mAb (148.3) or mouse IgG coupled to agarose beads (C=Ctrl), stripped, and reprecipitated with R.gp48C recognizing tapasin. Samples were incubated with DTT before SDS–PAGE where indicated. The tapasin/ERp57 conjugate, free ERp57, and free and reduced tapasin are indicated. The upper band in non-DTT-treated samples represents partially oxidized monomeric tapasin, and the lower band corresponds to fully oxidized tapasin. The lower panel shows quantitation of the partially oxidized upper band and the conjugate throughout the chase period.
Figure 5
Monoglucosylated _N_-linked glycans are not required for conjugate formation. (A) FO-1.β2m cells were incubated with or without 2 mM CST throughout starvation, pulse-labeling, and chase. All samples were treated as in Figure 3 and immunoprecipitated tapasin and its conjugate with ERp57 were subjected to SDS–PAGE without reduction. The right panel depicts the percent maximum conjugate signal throughout the chase period. (B) CNX is dispensable for conjugate formation. CNX-negative CEM.NKR cells expressing HLA-A2 or HLA-A2 and CNX were solubilized in Triton X-100 and post-nuclear supernatants were precipitated with M.PDI, PaSta1 (anti-tapasin mAb), or MaP.ERp57. SDS–PAGE was performed under reducing or nonreducing conditions, and, following transfer, membranes were probed for tapasin with R.gp48C. The tapasin/ERp57 conjugate and free tapasin are indicated.
Figure 6
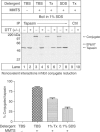
Noncovalent interactions prevent conjugate reduction. Noncovalent interactions within the MHC class I loading complex inhibit ERp57 escape pathway activation. HLA-A, -B, and -C-negative .221 cells were labeled with [35S]methionine and cysteine for 30 min and chased for an additional 30 min before membrane preparation. Membranes were resuspended in TBS, 1% Triton X-100, or 0.1% SDS in TBS with 1 mM CaCl2 for 1 h on ice before MMTS addition where indicated. All samples were then heated at 95°C for 5 min in 1% SDS, brought to 1 ml with 1% Triton X-100 in TBS with 2 mM MMTS, and immunoprecipitated with rabbit anti-tapasin (R.SinA) or normal rabbit serum and protein A Sepharose. SDS–PAGE was performed under nonreducing or reducing conditions as indicated, and percent conjugated tapasin was calculated. The tapasin/ERp57 conjugate, free ERp57, and free and reduced tapasin are indicated. Data shown are the average±s.e.m. of two independent experiments.
Figure 7
Tapasin alone stabilizes a mixed disulfide with ERp57. (A) The C60A mutant of ERp57 forms a mixed disulfide with native tapasin in vitro. Wild-type, C60A, and C409A mutant ERp57 molecules were mixed with soluble, recombinant tapasin in the presence of the indicated ratios of reduced (GSH) and oxidized (GSSG) glutathione. SDS sample buffer was added to all samples before SDS–PAGE. (B) In vitro formed conjugates contain both ERp57 and tapasin. Samples were treated as in (A), but after SDS–PAGE the samples were transferred to PVDF membranes and probed for tapasin with R.SinA or for ERp57 with a rabbit antiserum to the intact protein. (C) Noncovalent interactions between tapasin and ERp57 maintain the conjugate. Soluble recombinant ERp57 and tapasin were incubated alone or in combination as indicated. Samples were subsequently treated with 10 mM MMTS for 10 min at RT before or after the addition of urea (final concentration 8 M, right panel) or 10 × nonreducing SDS–PAGE sample buffer (left panel). Gels were run under nonreducing conditions and proteins detected by Coomassie Blue staining. The tapasin/ERp57 conjugate, free ERp57, and free tapasin are indicated. The conjugate composed of tapasin and WT ERp57 dissociates upon treatment with urea or SDS, but preincubation with MMTS or deletion of Cys-60 inhibits escape pathway activation and preserves the disulfide linkage.
Figure 8
Noncovalent interactions between tapasin and ERp57 prevent escape pathway activation. Under native conditions, noncovalent interactions between tapasin and ERp57 eliminate the ability of the sulfhydryl of Cys-60 of ERp57 to attack the mixed disulfide between tapasin and ERp57. We hypothesize that tapasin reduces the access of Cys-60 to the mixed disulfide and/or alters the local environment to prevent Cys-60 deprotonation. When denatured, the interactions of tapasin with ERp57 are disrupted, and the sulfhydryl of Cys-60 is now able to attack the mixed disulfide, leading to conjugate reduction. We speculate that the mixed disulfide with tapasin serves a structural rather than enzymatic function, and the altered redox state of MHC class I heavy chain in loading complexes deficient in ERp57 is likely due to the absence of the active site within the a′ domain of ERp57.
Similar articles
- Disulfide bond isomerization and the assembly of MHC class I-peptide complexes.
Dick TP, Bangia N, Peaper DR, Cresswell P. Dick TP, et al. Immunity. 2002 Jan;16(1):87-98. doi: 10.1016/s1074-7613(02)00263-7. Immunity. 2002. PMID: 11825568 - The thiol oxidoreductase ERp57 is a component of the MHC class I peptide-loading complex.
Hughes EA, Cresswell P. Hughes EA, et al. Curr Biol. 1998 Jun 4;8(12):709-12. doi: 10.1016/s0960-9822(98)70278-7. Curr Biol. 1998. PMID: 9637923 - Characterization of the ERp57-Tapasin complex by rapid cellular acidification and thiol modification.
Antoniou AN, Powis SJ. Antoniou AN, et al. Antioxid Redox Signal. 2003 Aug;5(4):375-9. doi: 10.1089/152308603768295104. Antioxid Redox Signal. 2003. PMID: 13678524 - Interaction of ERp57 and tapasin in the generation of MHC class I-peptide complexes.
Garbi N, Hämmerling G, Tanaka S. Garbi N, et al. Curr Opin Immunol. 2007 Feb;19(1):99-105. doi: 10.1016/j.coi.2006.11.013. Epub 2006 Dec 5. Curr Opin Immunol. 2007. PMID: 17150345 Review. - The optimization of peptide cargo bound to MHC class I molecules by the peptide-loading complex.
Elliott T, Williams A. Elliott T, et al. Immunol Rev. 2005 Oct;207:89-99. doi: 10.1111/j.0105-2896.2005.00311.x. Immunol Rev. 2005. PMID: 16181329 Review.
Cited by
- A personal retrospective on the mechanisms of antigen processing.
Cresswell P. Cresswell P. Immunogenetics. 2019 Mar;71(3):141-160. doi: 10.1007/s00251-018-01098-2. Epub 2019 Jan 29. Immunogenetics. 2019. PMID: 30694344 Free PMC article. Review. - Profiling thiol metabolites and quantification of cellular glutathione using FT-ICR-MS spectrometry.
Gori SS, Lorkiewicz P, Ehringer DS, Belshoff AC, Higashi RM, Fan TW, Nantz MH. Gori SS, et al. Anal Bioanal Chem. 2014 Jul;406(18):4371-9. doi: 10.1007/s00216-014-7810-z. Epub 2014 May 25. Anal Bioanal Chem. 2014. PMID: 24858467 Free PMC article. - Assembly of MHC class I molecules within the endoplasmic reticulum.
Zhang Y, Williams DB. Zhang Y, et al. Immunol Res. 2006;35(1-2):151-62. doi: 10.1385/IR:35:1:151. Immunol Res. 2006. PMID: 17003517 Review. - Human cytomegalovirus disrupts the major histocompatibility complex class I peptide-loading complex and inhibits tapasin gene transcription.
Halenius A, Hauka S, Dölken L, Stindt J, Reinhard H, Wiek C, Hanenberg H, Koszinowski UH, Momburg F, Hengel H. Halenius A, et al. J Virol. 2011 Apr;85(7):3473-85. doi: 10.1128/JVI.01923-10. Epub 2011 Jan 19. J Virol. 2011. PMID: 21248040 Free PMC article. - Disulfide bond formation in the mammalian endoplasmic reticulum.
Bulleid NJ. Bulleid NJ. Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a013219. doi: 10.1101/cshperspect.a013219. Cold Spring Harb Perspect Biol. 2012. PMID: 23125019 Free PMC article. Review.
References
- Antoniou AN, Powis SJ (2003) Characterization of the ERp57–Tapasin complex by rapid cellular acidification and thiol modification. Antioxid Redox Signal 5: 375–379 - PubMed
- Bangia N, Lehner PJ, Hughes EA, Surman M, Cresswell P (1999) The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur J Immunol 29: 1858–1870 - PubMed
- Braakman I, Helenius J, Helenius A (1992) Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature 356: 260–262 - PubMed
- Chen M, Stafford WF, Diedrich G, Khan A, Bouvier M (2002) A characterization of the lumenal region of human tapasin reveals the presence of two structural domains. Biochemistry 41: 14539–14545 - PubMed
- Cresswell P (2000) Intracellular surveillance: controlling the assembly of MHC class I–peptide complexes. Traffic 1: 301–305 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous