Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing different PPAR isoforms - PubMed (original) (raw)
doi: 10.1186/1478-1336-3-3.
Yumi Kobayashi, Toshiya Tanaka, Masayuki Tagami, Akira Sugiyama, Tatsuya Katayama, Chihiro Ueda, Daisuke Yamasaki, Kenji Ishimoto, Mikako Sumitomo, Yasutoshi Uchiyama, Takahide Kohro, Juro Sakai, Takao Hamakubo, Tatsuhiko Kodama, Takefumi Doi
Affiliations
- PMID: 16197558
- PMCID: PMC1262764
- DOI: 10.1186/1478-1336-3-3
Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing different PPAR isoforms
Keisuke Tachibana et al. Nucl Recept. 2005.
Abstract
Background: Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors and commonly play an important role in the regulation of lipid homeostasis. To identify human PPARs-responsive genes, we established tetracycline-regulated human hepatoblastoma cell lines that can be induced to express each human PPAR and investigated the gene expression profiles of these cells.
Results: The expression of each introduced PPAR gene was investigated using the various concentrations of doxycycline in the culture media. We found that the expression of each PPAR subtype was tightly controlled by the concentration of doxycycline in these established cell lines. DNA microarray analyses using these cell lines were performed with or without adding each subtype ligand and provided much important information on the PPAR target genes involved in lipid metabolism, transport, storage and other activities. Interestingly, it was noted that while ligand-activated PPARdelta induced target gene expression, unliganded PPARdelta repressed these genes. The real-time RT-PCR was used to verify the altered expression of selected genes by PPARs and we found that these genes were induced to express in the same pattern as detected in the microarray analyses. Furthermore, we analysed the 5'-flanking region of the human adipose differentiation-related protein (adrp) gene that responded to all subtypes of PPARs. From the detailed analyses by reporter assays, the EMSAs, and ChIP assays, we determined the functional PPRE of the human adrp gene.
Conclusion: The results suggest that these cell lines are important tools used to identify the human PPARs-responsive genes.
Figures
Figure 1
Induction of the expression of PPAR by doxycycline in established cell lines. A, B, C and D, Nuclear extracts (50 μg protein/lane) from each cell line cultured in the presence of the indicated amounts of Dox for 5 days were subjected to SDS-PAGE and immunoblots were performed with anti-PPARα (A), anti-PPARδ (B) or anti-PPARγ (C and D).
Figure 2
Time course of PPAR expression in established cell lines. A, B, C and D, The cell lines for the expression of PPARα (A), PPARδ (B), PPARγ1 (C) or PPARγ2 (D) were cultured in the presence of Dox. At time point 0, Dox was removed from the medium. The total RNAs were extracted from the cells cultured for the indicated days after removal of Dox and the amount of mRNAs of PPARs were measured by real time RT-PCR. Values are expressed as the mean ± S.E.M. (n = 3), relative to the control (0 day) set as 1. E, F, G and H, Nuclear extracts (50 μg protein/lane) from the cell lines expressing PPARα (E), PPARδ (F), PPARγ1 (G) or PPARγ2 (H) cultured for the indicated days after removal of Dox were subjected to SDS-PAGE and immunoblots were performed with anti-PPARα (E), anti-PPARδ (F) or anti-PPARγ (G and H).
Figure 3
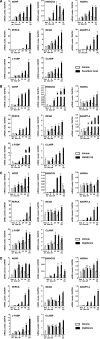
Eight gene expressions were modulated by PPARs in tet-regulated HepG2 cells. A, B, C and D, HepG2-tet-off-hPPAR cells were treated with DMSO (vehicle) or PPAR ligands (100 μM fenofibric acid for PPARα (A), 100 nM GW501516 for PPARδ (B), 10 μM ciglitizone for PPARγ1 (C) or PPARγ2 (D)) for 0, 8, 24, 48 or 72 h in the absence of Dox. Messenger RNA levels of human ADRP, HMGCS2, HADHA, PEPCK, MCAD, ANGPTL4, L-FABP and CLAMP/PDZK1 were measured by real time RT-PCR. Values are expressed as mean ± S.E.M. (n = 3) target mRNA levels normalized to GAPDH mRNA.
Figure 4
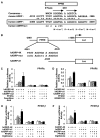
PPARs modulate human ADRP promoter activity via a PPRE located between -2361 and -2345 bp. A, A sequence corresponding to the -2366/-2339 region of the human adrp gene, was compared with a consensus PPRE and the analogous regions in the mouse adrp gene promoter (-2013/-1986). A human ADRPmut indicates a human ADRP promoter whose potential PPRE was mutated. Asterisks denote conserved bases and arrows represent response element half-sites. B, Schematic representation of the chimeric genes containing the human ADRP promoters; each wild type (hADRP-4K), point mutation (hADRP-mut), and deletion (hADRP-d1) of the ADRP promoter was cloned in front of the firefly luciferase reporter gene. Lowercase letters indicate mutations introduced in the human ADRP PPRE. C, D, E and F, HepG2 cells were co-transfected with a human ADRP reporter plasmid (50 ng), phRL-TK (50 ng) and either pcDNA3-hPPARα (5 ng) (C), pcDNA3-hPPARδ (5 ng) (D), pcDNA3-hPPARγ1 (5 ng) (E) or pcDNA3-hPPARγ2 (5 ng) (F). Transfected cells were treated with ligands (100 μM fenofibric acid (C), 100 nM GW501516 (D) or 10 μM ciglitizone (E and F)) for 24 h and the cells were used for reporter gene assays. Luciferase activities from reporter plasmids were normalized by internal Renilla luciferase activity. Values are expressed as fold induction of the control (the value when only reporter plasmid (ADRP-4K) was transfected) set at 1. Values represent the mean ± S.E.M. (n = 3).
Figure 5
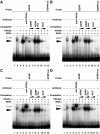
PPARs bind to the PPRE in the -2366/-2339 region of the human adrp gene. A, B, C and D, EMSAs were performed with 32P-labelled either ADRP or mutated ADRP (ADRPmut) oligonucleotides in the presence of purified PPARα (A), purified PPARδ (B), in vitro transcribed/translated PPARγ1 (C), purified PPARγ2 (D) and/or purified RXRα proteins. Supershift experiments were carried out using anti-PPARα (A), anti-PPARδ (B) or anti-PPARγ (C and D) antibodies. Unlabelled oligonucleotides (ADRP, ADRPmut or ACO) were used at 10- or 100-fold molar excess to the labelled probe to perform competition assays. Closed and open arrowheads indicate the specific bands and the supershift bands, respectively.
Figure 6
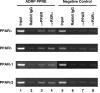
Chromatin immunoprecipitation assays of the ADRP promoter. HepG2-tet-off-hPPAR cells were treated with PPAR ligands (100 μM fenofibric acid for PPARα, 100 nM GW501516 for PPARδ, or 10 μM troglitazone for PPARγ) for 8 h in the absence of Dox and processed for the ChIP assays. Soluble chromatin was immunoprecipitated with pre-immune rabbit IgG (lanes 2 and 6), anti-PPAR antibodies (lanes 3 and 7), or anti-RXRα antibody (lanes 4 and 8). Immunoprecipitates were subjected to PCR with a primer-pair specific to the ADRP promoter. As a negative control, a second set of primers were used to amplify another genomic region that was not expected to interact with the PPARs. PCR was performed with total chromatin input (lanes 1 and 5).
Similar articles
- [Application of the human hepatoblastoma cell lines inducibly expressing peroxisome proliferator-activated receptors (PPARs)].
Tachibana K. Tachibana K. Yakugaku Zasshi. 2007 Aug;127(8):1223-9. doi: 10.1248/yakushi.127.1223. Yakugaku Zasshi. 2007. PMID: 17666873 Review. Japanese. - The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis.
Hummasti S, Tontonoz P. Hummasti S, et al. Mol Endocrinol. 2006 Jun;20(6):1261-75. doi: 10.1210/me.2006-0025. Epub 2006 Mar 23. Mol Endocrinol. 2006. PMID: 16556736 - PPAR: a mediator of peroxisome proliferator action.
Green S. Green S. Mutat Res. 1995 Dec;333(1-2):101-9. doi: 10.1016/0027-5107(95)00136-0. Mutat Res. 1995. PMID: 8538617 Review. - Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression.
Marcus SL, Miyata KS, Zhang B, Subramani S, Rachubinski RA, Capone JP. Marcus SL, et al. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5723-7. doi: 10.1073/pnas.90.12.5723. Proc Natl Acad Sci U S A. 1993. PMID: 8390676 Free PMC article. - Peroxisome proliferators and peroxisome proliferator activated receptors (PPARs) as regulators of lipid metabolism.
Latruffe N, Vamecq J. Latruffe N, et al. Biochimie. 1997 Feb-Mar;79(2-3):81-94. doi: 10.1016/s0300-9084(97)81496-4. Biochimie. 1997. PMID: 9209701 Review.
Cited by
- Cellular and molecular consequences of peroxisome proliferator-activated receptor-gamma activation in ovarian cancer cells.
Vignati S, Albertini V, Rinaldi A, Kwee I, Riva C, Oldrini R, Capella C, Bertoni F, Carbone GM, Catapano CV. Vignati S, et al. Neoplasia. 2006 Oct;8(10):851-61. doi: 10.1593/neo.06433. Neoplasia. 2006. PMID: 17032502 Free PMC article. - The Role of PPARs in Cancer.
Tachibana K, Yamasaki D, Ishimoto K, Doi T. Tachibana K, et al. PPAR Res. 2008;2008:102737. doi: 10.1155/2008/102737. PPAR Res. 2008. PMID: 18584037 Free PMC article. - The PPARγ-SETD8 axis constitutes an epigenetic, p53-independent checkpoint on p21-mediated cellular senescence.
Shih CT, Chang YF, Chen YT, Ma CP, Chen HW, Yang CC, Lu JC, Tsai YS, Chen HC, Tan BC. Shih CT, et al. Aging Cell. 2017 Aug;16(4):797-813. doi: 10.1111/acel.12607. Epub 2017 May 17. Aging Cell. 2017. PMID: 28514051 Free PMC article. - Pharmacological correction of a defect in PPAR-gamma signaling ameliorates disease severity in Cftr-deficient mice.
Harmon GS, Dumlao DS, Ng DT, Barrett KE, Dennis EA, Dong H, Glass CK. Harmon GS, et al. Nat Med. 2010 Mar;16(3):313-8. doi: 10.1038/nm.2101. Epub 2010 Feb 14. Nat Med. 2010. PMID: 20154695 Free PMC article. - Integration site choice of a feline immunodeficiency virus vector.
Kang Y, Moressi CJ, Scheetz TE, Xie L, Tran DT, Casavant TL, Ak P, Benham CJ, Davidson BL, McCray PB Jr. Kang Y, et al. J Virol. 2006 Sep;80(17):8820-3. doi: 10.1128/JVI.00719-06. J Virol. 2006. PMID: 16912328 Free PMC article.
References
- Corton JC, Lapinskas PJ, Gonzalez FJ. Central role of PPARalpha in the mechanism of action of hepatocarcinogenic peroxisome proliferators. Mutat Res. 2000;448:139–151. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials