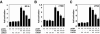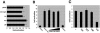LMCD1/Dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding - PubMed (original) (raw)
LMCD1/Dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding
Nibedita Rath et al. Mol Cell Biol. 2005 Oct.
Abstract
The activity of GATA factors is regulated, in part, at the level of protein-protein interactions. LIM domain proteins, first defined by the zinc finger motifs found in the Lin11, Isl-1, and Mec-3 proteins, act as coactivators of GATA function in both hematopoietic and cardiovascular tissues. We have identified a novel GATA-LIM interaction between GATA6 and LMCD1/dyxin. The LIM domains and cysteine-rich domains in LMCD1/dyxin and the carboxy-terminal zinc finger of GATA6 mediate this interaction. Expression of LMCD1/dyxin is remarkably similar to that of GATA6, with high-level expression observed in distal airway epithelium of the lung, vascular smooth muscle, and myocardium. In contrast to other GATA-LIM protein interactions, LMCD1/dyxin represses GATA6 activation of both lung and cardiac tissue-specific promoters. Electrophoretic mobility shift and chromatin immunoprecipitation assays show that LMCD1/dyxin represses GATA6 function by inhibiting GATA6 DNA binding. These data reveal an interaction between GATA6 and LMCD1/dyxin and demonstrate a novel mechanism through which LIM proteins can assert their role as transcriptional cofactors of GATA proteins.
Figures
FIG. 1.
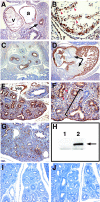
Identification of LMCD1/dyxin as a GATA6 binding protein. (A) The Cytotrap yeast two-hybrid screening strategy. The bait protein, in this case GATA6, is expressed in frame with human Sos. Target proteins in the library are expressed with a myristoylation sequence, causing them to locate on the inner side of the plasma membrane. Interaction between bait and target proteins colocalizes Sos with Ras, which activates the Ras signal transduction pathway, allowing growth of yeast strain cdc25H on galactose at 37°C. (B) Screening of a human adult lung cDNA library with GATA6 as the bait protein resulted in the identification of three clones homologous to human LMCD1/dyxin. LMCD1/dyxin has a conserved cysteine-rich domain at the amino-terminal end (blue), two LIM domains (red), and a putative nuclear localization motif (green). The amino-terminal end of the proteins encoded by the partial cDNAs isolated is noted by black arrowheads. (C) The LMCD1/dyxin cDNA clone encoding aa 91 to 365 was retransformed into the cdc25H yeast strain with blank pSOS plasmid or with pSOS/GATA6. Activation of the Ras signal transduction pathway, which results in the ability of the cdc25H yeast strain to grow at 37°C on galactose, occurred only with coexpression of GATA6.
FIG. 2.
Protein-protein interaction between GATA6 and LMCD1/dyxin. (A) GST pull-down assay showing the interaction between GATA6 and full-length LMCD1/dyxin. (B) Coimmunoprecipitation of GATA6 and LMCD1 from MLE-15 cell extracts. Extracts were immunoprecipitated with a GATA6 antibody and then analyzed by Western blotting with the LMCD1/dyxin polyclonal antibody. “Input” represents 5% of the extract used in the immunoprecipitation. Position of the 35-kDa molecular mass marker is shown. Expression of GATA6 in MLE-15 cells has already been reported (6, 31). IP, immunoprecipitation; WB, Western blot; NIgG, normal IgG; G6 Ab, GATA6 antibody. (C) GST pull-down assay showing interaction between the LIM and CYS domains and GATA6. Arrowheads in panels A and C indicate radiolabeled GATA6 interaction with GST-LMCD1 fusion proteins. (D) Coomassie blue-stained gel of GST fusion proteins. Asterisks denote full-length fusion proteins. Molecular mass standards in kilodaltons are noted on the left. (E) Far-Western blot showing interaction between GATA6 and the LIM- and cysteine-rich domains of LMCD1/dyxin. The arrow and asterisks denote GATA6 interaction with GST fusion proteins. The arrowhead denotes a nonspecific band that reacts with anti-GST-LMCD1 antisera. Mock-transfected cell extracts were probed with full-length GST-LMCD1. Molecular mass standards in kilodaltons are noted on the left. (F) Summary of the domains within LMCD1/dyxin that interact with GATA6. −, no interaction; +++, interaction. (G) GST pull-down assay to assess the abilities of different regions of GATA6 that interact with LMCD1/dyxin, as well as full-length GATA1 and GATA4. Molecular mass standards in kilodaltons are noted on the left. (H) Input of radiolabeled GATA6, GATA1, and GATA4 proteins. Molecular mass standards in kilodaltons are noted on the left. (I) Summary of domains within GATA6 that interact with LMCD1/dyxin. These regions delineate the carboxy-terminal zinc finger as the domain in GATA6 required for interaction with LMCD1/dyxin. Gel lane numbers in panels G and H correspond to the GATA6 proteins indicated in panel I.
FIG. 3.
LMCD1/dyxin gene expression during cardiopulmonary development. In situ hybridization was performed with tissue sections from E12.5 (A, B, G, and H), E14.5 (C and D), and E18.5 (E and F) mouse embryos. Expression is observed in developing airway epithelium (A, arrows) with the highest expression at the distal tips of the airways (B, arrowheads). Expression is also observed in the underlying mesenchyme (A). Expression continues in the lung airways at E14.5 (C) but decreases by E18.5 so that it is observed in only a small subset of alveolar epithelium (F, arrowheads). Expression in the cardiovascular system is observed in the muscular component of the dorsal aorta (D) and in the pulmonary arteries (E). Expression in the heart (H) is observed in the myocardium as well as the developing endocardial cushions (arrows). ao, aorta; ua, upper airways; eso, esophagus; pa, pulmonary artery.
FIG. 4.
Expression of LMCD1/dyxin protein during cardiopulmonary development. Immunohistochemistry was performed with tissue sections from E9.5 (A and B), E12.5 (C and D), E14.5 (E, F, I, and J), and E16.5 (G) mouse embryos. LMCD1/dyxin protein expression was observed in the developing heart at E9.5 (A and B) and E12.5 (D). LMCD1/dyxin expression is observed in the anterior foregut endoderm at E9.5 (A, arrow). In contrast to gene expression, LMCD1/dyxin protein expression is not observed in the developing endocardial cushions of the heart at E12.5 (D, arrows). Expression is observed throughout the branching airways of the lung at E12.5 (C). By E14.5, LMCD1/dyxin expression is found in the distal airway epithelium in the lung (E), with the highest levels observed in the distal regions (F, bracket). Expression is also observed in the developing pulmonary arteries (E). In both heart and lung sections, note the presence of nuclear LMCD1/dyxin expression in a subset of cells (B and F, arrowheads). The anti-GST-LMCD1 antibody recognizes LMCD1/dyxin protein from transfected (lane 2) but not untransfected (lane 1) 293 cells (H, arrow). Use of preimmune serum (I) and preincubation of LMCD1 antiserum with LMCD1 fusion protein (J) results in loss of staining in cardiopulmonary tissues. v, ventricle; a, atrium; ves, pulmonary vessel.
FIG. 5.
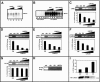
LMCD1/dyxin is retained in the nucleus of leptomycin B-treated cells. 293 cells were transfected with a myc-tagged LMCD1 expression construct and treated with 20 μM leptomycin B for 4 hours. Cells were stained with the anti-myc epitope antibody (9E10). Untreated cells showed primarily cytoplasmic staining of LMCD1/dyxin (A), whereas leptomycin B-treated cells showed an accumulation of protein within the nucleus. (B) Fluorescein isothiocyanate staining of LMCD1; (C) DAPI (4′,6′-diamidino-2-phenylindole) staining; (D) overlay of panels B and C. Coexpression of LMCD1 (E) and GATA6 (F) does not change cellular localization of GATA6. Panel G is an overlay of panels E and F.
FIG. 6.
LMCD1/dyxin represses GATA6-mediated trans activation of lung- and cardiac tissue-specific promoters. NIH 3T3 cells were transfected with the indicated amounts (in micrograms) of GATA6 and LMCD1/dyxin expression plasmids or empty vector along with the mouse SP-A luciferase reporter plasmid 150 bp upstream of the transcriptional start site (A) or the −124-bp cTNC growth hormone reporter plasmid (B). An identical assay was performed with GATA4 and the cTNC promoter (C). Data represent the averages of results of three assays ± standard errors of the means.
FIG. 7.
LMCD1/dyxin does not have inherent transcriptional repression activity, and the full-length protein is required for repression. LMCD1/dyxin, or domains within, does not repress a GAL4-responsive reporter containing the simian virus 40 promoter (A). A GAL4-GATA6 fusion protein was used to activate the GAL4 response reporter, and increasing amounts of LMCD1 were coexpressed. LMCD1/dyxin does not affect the activity of GATA6 when fused to the GAL4 DNA binding domain (B). The indicated domains in LMCD1/dyxin were expressed, along with GATA6 and the SP-A luciferase reporter in 293 cells, to show that full-length LMCD1/dyxin can repress GATA6 activity on the SP-A promoter but that individual domains within LMCD1/dyxin cannot (C). Data represent the averages of results of three assays ± standard errors of the means.
FIG. 8.
LMCD1/dyxin inhibits GATA6 DNA binding and represses GATA6 function in vivo. Gel shift assays were used to assess the ability of LMCD1/dyxin to affect GATA6 DNA binding. In vitro-translated GATA6 binds to the conserved GATA sites within the mouse Wnt7b promoter (A, lanes 1 and 4). Increasing expression of LMCD1/dyxin inhibits GATA6 DNA binding when LMCD1/dyxin is cotranslated in an in vitro transcription/translation reaction (A, lanes 5 and 6), while increasing amounts of an empty plasmid did not have any effect (A, lanes 2 and 3). Addition of increasing amounts of GST-LMCD1 (B, lanes 10 to 12), but not GST alone (B, lanes 6 to 8), inhibits GATA6 DNA binding. Note that GST-LMCD1 does not bind to DNA (B, lanes 1 to 4). ChIP assays show that increasing levels of LMCD1/dyxin inhibit GATA6 DNA binding to conserved sites located in the cTNC (C) and SP-C (D) promoters as well as that of GATA1 (E) and GATA4 (F) to the cTNC promoter. However, LMCD1/dyxin does not inhibit Foxp4 binding to the forkhead DNA binding site located in the mouse CC10 promoter (G). Q-PCR ChIP data are shown as a percentage of GATA or Foxp4 binding without LMCD1. GATA6 but not LMCD1/dyxin is found associated with the conserved GATA DNA binding site located in the mouse SP-C promoter (H). siRNA transfection of MLE-15 cells with the indicated amounts of oligonucleotides (in micrograms) resulted in inhibition of LMCD1/dyxin expression (indicated in Western blot in bottom panel) and an increase in SP-A expression (I). The position of the 35-kDa molecular mass marker is shown in panel I. IVT, in vitro translated; ext, extract; −Ab, without antibody; G6, GATA6; G1, GATA1; G4, GATA4; P4, Foxp4; WT, wild type; WB, Western blot.
FIG. 9.
DNA binding inhibition by LMCD1/dyxin defines a new mechanism to restrict GATA factor function. (A) LMCD1/dyxin represses GATA6 function through inhibition of DNA binding. Strict control over cellular localization of LMCD1/dyxin may define its regulatory function. (B) GATA factors also interact with a variety of corepressors, such as FOG-1/2. These interactions repress GATA function by recruiting repression function in trans, sometimes through interaction with additional corepressor proteins such as CtBP-1 (8, 15, 34). Both models effectively restrict GATA factor activity.
Similar articles
- Lmcd1/Dyxin, a novel Z-disc associated LIM protein, mediates cardiac hypertrophy in vitro and in vivo.
Frank D, Frauen R, Hanselmann C, Kuhn C, Will R, Gantenberg J, Füzesi L, Katus HA, Frey N. Frank D, et al. J Mol Cell Cardiol. 2010 Oct;49(4):673-82. doi: 10.1016/j.yjmcc.2010.06.009. Epub 2010 Jun 30. J Mol Cell Cardiol. 2010. PMID: 20600098 - A novel p38 MAPK target dyxin is rapidly induced by mechanical load in the heart.
Luosujärvi H, Aro J, Tokola H, Leskinen H, Tenhunen O, Skoumal R, Szokodi I, Ruskoaho H, Rysä J. Luosujärvi H, et al. Blood Press. 2010 Feb;19(1):54-63. doi: 10.3109/08037050903464519. Blood Press. 2010. PMID: 20175653 - LIM and cysteine-rich domains 1 (LMCD1) regulates skeletal muscle hypertrophy, calcium handling, and force.
Ferreira DMS, Cheng AJ, Agudelo LZ, Cervenka I, Chaillou T, Correia JC, Porsmyr-Palmertz M, Izadi M, Hansson A, Martínez-Redondo V, Valente-Silva P, Pettersson-Klein AT, Estall JL, Robinson MM, Nair KS, Lanner JT, Ruas JL. Ferreira DMS, et al. Skelet Muscle. 2019 Oct 31;9(1):26. doi: 10.1186/s13395-019-0214-1. Skelet Muscle. 2019. PMID: 31666122 Free PMC article. - Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2.
Hill AA, Riley PR. Hill AA, et al. Mol Cell Biol. 2004 Nov;24(22):9835-47. doi: 10.1128/MCB.24.22.9835-9847.2004. Mol Cell Biol. 2004. PMID: 15509787 Free PMC article. - Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins.
Cantor AB, Orkin SH. Cantor AB, et al. Semin Cell Dev Biol. 2005 Feb;16(1):117-28. doi: 10.1016/j.semcdb.2004.10.006. Epub 2004 Dec 15. Semin Cell Dev Biol. 2005. PMID: 15659346 Review.
Cited by
- Harnessing conserved signaling and metabolic pathways to enhance the maturation of functional engineered tissues.
Callaghan NI, Durland LJ, Ireland RG, Santerre JP, Simmons CA, Davenport Huyer L. Callaghan NI, et al. NPJ Regen Med. 2022 Sep 3;7(1):44. doi: 10.1038/s41536-022-00246-3. NPJ Regen Med. 2022. PMID: 36057642 Free PMC article. Review. - Proteomics Recapitulates Ovarian Proteins Relevant to Puberty and Fertility in Brahman Heifers (Bos indicus L.).
Tahir MS, Nguyen LT, Schulz BL, Boe-Hansen GA, Thomas MG, Moore SS, Lau LY, Fortes MRS. Tahir MS, et al. Genes (Basel). 2019 Nov 12;10(11):923. doi: 10.3390/genes10110923. Genes (Basel). 2019. PMID: 31726744 Free PMC article. - LIM and cysteine-rich domains 1 is required for thrombin-induced smooth muscle cell proliferation and promotes atherogenesis.
Janjanam J, Zhang B, Mani AM, Singh NK, Traylor JG Jr, Orr AW, Rao GN. Janjanam J, et al. J Biol Chem. 2018 Mar 2;293(9):3088-3103. doi: 10.1074/jbc.RA117.000866. Epub 2018 Jan 11. J Biol Chem. 2018. PMID: 29326163 Free PMC article. - Genetic association analyses highlight biological pathways underlying mitral valve prolapse.
Dina C, Bouatia-Naji N, Tucker N, Delling FN, Toomer K, Durst R, Perrocheau M, Fernandez-Friera L, Solis J; PROMESA investigators; Le Tourneau T, Chen MH, Probst V, Bosse Y, Pibarot P, Zelenika D, Lathrop M, Hercberg S, Roussel R, Benjamin EJ, Bonnet F, Lo SH, Dolmatova E, Simonet F, Lecointe S, Kyndt F, Redon R, Le Marec H, Froguel P, Ellinor PT, Vasan RS, Bruneval P, Markwald RR, Norris RA, Milan DJ, Slaugenhaupt SA, Levine RA, Schott JJ, Hagege AA; MVP-France; Jeunemaitre X; Leducq Transatlantic MITRAL Network. Dina C, et al. Nat Genet. 2015 Oct;47(10):1206-11. doi: 10.1038/ng.3383. Epub 2015 Aug 24. Nat Genet. 2015. PMID: 26301497 Free PMC article. - Human pre-valvular endocardial cells derived from pluripotent stem cells recapitulate cardiac pathophysiological valvulogenesis.
Neri T, Hiriart E, van Vliet PP, Faure E, Norris RA, Farhat B, Jagla B, Lefrancois J, Sugi Y, Moore-Morris T, Zaffran S, Faustino RS, Zambon AC, Desvignes JP, Salgado D, Levine RA, de la Pompa JL, Terzic A, Evans SM, Markwald R, Pucéat M. Neri T, et al. Nat Commun. 2019 Apr 26;10(1):1929. doi: 10.1038/s41467-019-09459-5. Nat Commun. 2019. PMID: 31028265 Free PMC article.
References
- Bekman, E., and D. Henrique. 2002. Embryonic expression of three mouse genes with homology to the Drosophila melanogaster prickle gene. Mech. Dev. 119(Suppl. 1):S77-S81. - PubMed
- Bespalova, I. N., and M. Burmeister. 2000. Identification of a novel LIM domain gene, LMCD1, and chromosomal localization in human and mouse. Genomics 63:69-74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases