Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor - PubMed (original) (raw)
. 2005 Oct;25(20):8938-47.
doi: 10.1128/MCB.25.20.8938-8947.2005.
Hitoshi Shimano, Masanori Nakakuki, Takashi Matsuzaka, Yoshimi Nakagawa, Takashi Yamamoto, Ryuichiro Sato, Akimitsu Takahashi, Hirohito Sone, Naoya Yahagi, Hiroaki Suzuki, Hideo Toyoshima, Nobuhiro Yamada
Affiliations
- PMID: 16199872
- PMCID: PMC1265776
- DOI: 10.1128/MCB.25.20.8938-8947.2005
Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor
Noriyuki Inoue et al. Mol Cell Biol. 2005 Oct.
Abstract
Sterol regulatory element-binding proteins (SREBPs) are membrane-bound transcription factors that regulate lipid synthetic genes. In contrast to SREBP-2, which regulates cellular cholesterol level in normal cells, SREBP-1a is highly expressed in actively growing cells and activates entire programs of genes involved in lipid synthesis such as cholesterol, fatty acids, triglycerides, and phospholipids. Previously, the physiological relevance of this potent activity of SREBP-1a has been thought to regulate the supply of membrane lipids in response to cell growth. Here we show that nuclear SREBP-1a and SREBP-2 bind directly to a novel SREBP binding site in the promoter of the p21(WAF1/CIP1) gene, the major cyclin-dependent kinase inhibitor, and strongly activate its promoter activity. Only the SREBP-1a isoform consistently causes induction of p21 at both the mRNA and protein levels. Colony formation assays and polyploidy of livers from transgenic mice suggest that activation of p21 by SREBP-1a could inhibit cell growth. Activation of endogenous SREBPs in lipid deprivation conditions was associated with induction of p21 mRNA and protein. Expression of p21 was reduced in SREBP-1 null mice. These data suggest a physiological role of SREBP-1a in p21 regulation. Identification of p21 as a new SREBP target might implicate a new paradigm in the link between lipid synthesis and cell growth.
Figures
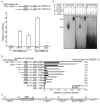
FIG. 1.
Activation of p21 gene promoter by SREBP-1a. a. Activation of p21 promoter-luciferase by SREBP-1a and p53. Saos-2 cells, p53-deficient osteosarcoma cell line, were transfected with the human p21 promoter firefly luciferase plasmid (p21-2338-Luc or p21-1363-Luc), expression vector for nuclear SREBP-1a or p53, and a Renilla luciferase reference plasmid, pRL-SV40. The luciferase activity was measured and normalized to the Renilla luciferase activity after 24 h incubation. p21-2338-Luc contains and p21-1363-Luc lacks two p53-binding sites. b. Identification of an SREBP binding site (SRE) in p21 gene promoter. A series of deletion luciferase constructs for the human p21-promoter are indicated in the left panel. HepG2 cells, human hepatocellular carcinoma cell line, were transfected with the indicated reporter plasmid, a cytomegalovirus expression plasmid for SREBP-1a or empty plasmid, and a reference plasmid, pRL-SV40. Relative luciferase activity was measured after 24 h incubation. Fold activation represents ratio of values for SREBP-1a versus a cytomegalovirus empty vector. c. The SREBP binding site (p21-SRE) and its neighboring region in human p21 gene promoter. d. SREBP-1a directly binds to p21-SRE. Gel mobility shift assays was performed to show the direct binding of SREBP-1a to the p21 promoter. Double-stranded DNA fragments corresponding to the LDLR-SRE (1 to 4), the original binding site for SREBP, p21-SRE (5 to 8), and p21-SRE-Mut (9 to 12) where the GT
GGGCCGA
G was replaced by GT
ACAAAAT
G, labeled with [α-32P]dCTP and incubated in the reaction mixture with (lanes 2 to 4, 6 to 8, and 10 to 12) or without (lane 1, 5, and 9) in vitro synthesized nuclear SREBP-1a protein. Specificity of SREBP-1a binding to the p21 SRE probe was confirmed by a supershift after the addition of SREBP-1 antibody (lane 8). In competition assays (Competitor), a 50-fold molar excess of an unlabeled SRE complex DNA (lane 7) was added to the incubation. RL, reticulocyte lysate; 1a, SREBP-1a.
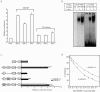
FIG. 2.
Binding characteristics of SREBP isoforms to the p21 gene promoter. a. Effects of SREBP isoforms and YR mutants on the p21 promoter. Luciferase assays for the p21-2338-Luc were compared among nuclear SREBP isoforms, and their respective YR mutants (SREBP-1aM −1 cM and −2 M), in which the tyrosine residue in the basic region of each SREBP isoform was replaced by arginine. These mutants were capable to bind to E-boxes as an innate feature of basic helix-loop-helx protein, but not to the SRE sites, SREBP-specific binding sites (12). cont, cytomegalovirus empty vector; BP1a, SREBP-1a; BP1c, SREBP-1c; BP2, SREBP-2. b. Gel mobility shift assay of nuclear SREBP-1 and -2. A double-stranded DNA fragment corresponding to p21-SRE was labeled with [α-32P]dCTP and incubated in the reaction mixture with (lanes 2, 3, 5, and 6) or without (lanes 1 and 4) in vitro synthesized nuclear SREBP-1a (1 to 3) or -2 (4 to 6) protein. Since SREBP-1a and -1c share the common basic helix-loop-helix domain, these data indicate that all the SREBP isoforms bind to the p21-SRE with similar affinities. c. The p21-SRE has an enhancer activity in the activation of p21 gene promoter by SREBPs. An SV40 promoter luciferase reporter for estimation of a region containing the upstream Sp1 site and p21-SRE as an enhancer was constructed as indicated. Deletion and mutation analysis were performed with reporter genes in the indicated positions of the promoter. HepG2 cells were transfected with each reporter plasmid, an expression plasmid for nuclear SREBP-1a or SREBP-2, reference plasmid pRL-SV40. Luciferase activity was measured and normalized to pRL-SV40 activity after 24 h incubation. Fold induction of luciferase activity by SREBPs is shown. d. Deletion of transactivation domain of SREBP-1 suppresses activation of p21 promoter by SREBP-1a and SREBP-2 in dominant negative fashion. SREBP-1 dominant negative form (DN-SREBP-1) lacks the amino-terminal transactivation domain (89 amino acids) and has the ability to bind to SRE site but inhibits transcriptional activities by native SREBPs for known SREBP target promoters (23). HepG2 cells were transfected with the p21-2338-Luc, SREBP expression vector (SREBP-1a or -2), and the indicated DNA amount of the dominant-negative SREBP-1 expression vector. Luciferase activity was measured after 24 h incubation.
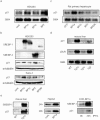
FIG. 3.
SREBP-1a induces p21 at mRNA and protein levels. a. p21 mRNA induction by overexpression of SREBPs in HEK293 cells. HEK293 cells, human kidney cell line were transfected with the indicated cytomegalovirus promoter expression plasmids. Total RNA was isolated from cells 24 h after transfection for Northern blot analysis (10 μg per lane). cont, cytomegalovirus empty vector; BP1a, SREBP-1a; BP1cm, SREBP-1c; BP2, SREBP-2. b. p21 protein induction by overexpression of SREBPs in HEK293 and Saos-2 cells. The cells were transfected with indicated cytomegalovirus promoter expression plasmid. Total cellular proteins from cells, 24 h after transfection of indicated expression plasmids. The protein was subjected to immunoblot analysis for SREBP-1 and SREBP-2 and p21 (42.5 μg per lane). c. p21 mRNA induction by overexpression of SREBPs in hepatocytes. Rat primary hepatocytes were infected with control adenovirus expressing GFP or an adenoviral vector expressing SREBPs at an MOI of 100. Total RNA was isolated from cells 48 h after infection for Northern blot analysis (10 μg per lane). GFP, green fluorescent protein for controls; DNBP, dominant-negative form of SREBP-1 described in the legend to Fig. 2d. d. p21 gene expression in livers from SREBP transgenic mice. Mice overexpressing a nuclear active form of human SREBP-1a, SREBP-1c, and SREBP-2 in the liver were fed a high-protein/low-carbohydrate diet for a week and fasted overnight to induce the transgene under the phosphoenolpyruvate carboxykinase (PEPCK) promoter. Total liver RNAs were subjected to Northern blot analysis to estimate gene expression with the indicated cDNA probes. WT, wild-type; BP1a, SREBP-1a transgenic mice; BP1c, SREBP-1c transgenic mice; BP2, SREBP-2 transgenic mice. e. Cellular p21 protein level in SREBP-1a overexpression. In the left panel, nuclear extract proteins from livers of wild-type or SREBP-1a transgenic mice were subjected to immunoblot analysis with antibody against SREBP-1 and p21. In the center panel, total cellular proteins from HepG2, 24 or 48 h after transfection of SREBP-1a expression plasmid, were subjected to immunoblot analysis with antibody against p21 (40 μg per lane). In the right panel, CHO stable cell lines that inducibly express nuclear SREBPs under IPTG-regulated promoter (18) were incubated in 0.1 mM IPTG-containing medium. Each cell was harvested in NP-40 buffer at the indicated time. Proteins were subjected to immunoblot analysis for SREBP-1 and p21 (100 μg per lane).
FIG. 4.
Induction of p21 by SREBP-1a leads to cell growth arrest as evidenced by colony formation assays with Saos-2 cells (a) and hepatocyte polyploidy of SREBP-1a transgenic and p21 null mice (b). a. Saos-2 cells were transfected with 10 μg of the indicated expression plasmids containing neomycin resistance gene; 24 h after transfection, the cells were cultured in medium containing 500 μg/ml of G418 for 3 weeks. The resultant survival cell colonies were fixed and stained with violet blue as representatively indicated in the upper panel. The values represent relative colony numbers normalized to an empty vector control. b. The hepatocytes were prepared for livers from the indicated mouse lines. The DNA content was determined by flow cytometry after DNA staining with propidium iodide. Hepatocyte ploidy is shown as 2n, 4n, and 8n. WT, C57BL/6J; Tg1a, SREBP-1a transgenic mice; p21KO, p21 knockout mice; Tg1a×p21KO, SREBP-1a transgenic and p21 knockout double mutant mice. **, P < 0.01, and *, P < 0.05 versus wild-type; #, P < 0.05 versus SREBP-1a transgenic mice (Student's t test).
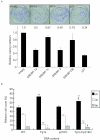
FIG. 5.
Activation of p21 promoter and induction of p21 protein by endogenous SREBPs in Saos-2 cells. Saos-2 cells were transfected with each reporter plasmid, SRE-Luc (a and c) or p21-2338-Luc (b and d), a reference plasmid pRL-CMV, and incubated at the indicated conditions to activate endogenous SREBPs by delipidated serum (a to d) and further pravastatin, an HMGCoA reductase inhibitor (e to h). Luciferase activity was measured and normalized to pRL-CMV activity after 24 h incubation (a, b, e, and f). Total cellular (40 μg) (c and d) and nuclear (10 μg) (g and h) proteins were subjected to immunoblot analysis for SREBP-1 and SREBP-2 (c and g), and p21 (d and h). FCS, 5% fetal calf serum; DLS, 5% delipidated serum; statin, 50 μM pravastatin; sterols, 10 μg/ml cholesterol and 1 μg/ml 25-hydroxychoresterol.
FIG. 6.
Disruption of endogenous SREBPs suppressed p21 expression. a. Saos-2 cells infected with control adenovirus expressing GFP or an adenoviral vector expressing dominant-negative SREBP-1 at a multiplicity of infection of 30. Total cellular proteins were isolated from cells incubated at the indicated conditions 24 h after infection. Total proteins (35 μg) were subjected to immunoblot analysis for SREBP-1, SREBP-2, and p21. b. Northern blot analysis was performed to measure p21 mRNA in livers and white adipose tissue (WAT) from wild-type (WT) and SREBP1 knockout (BPKO) mice. The animals were refed a high-sucrose/fat-free diet for 12 h after 24 h of starvation.
Similar articles
- A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G(1) cell-cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors.
Nakakuki M, Shimano H, Inoue N, Tamura M, Matsuzaka T, Nakagawa Y, Yahagi N, Toyoshima H, Sato R, Yamada N. Nakakuki M, et al. FEBS J. 2007 Sep;274(17):4440-52. doi: 10.1111/j.1742-4658.2007.05973.x. Epub 2007 Jul 27. FEBS J. 2007. PMID: 17662109 - SREBP transcription factors: master regulators of lipid homeostasis.
Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. Eberlé D, et al. Biochimie. 2004 Nov;86(11):839-48. doi: 10.1016/j.biochi.2004.09.018. Biochimie. 2004. PMID: 15589694 Review. - Acetyl-coenzyme A synthetase is a lipogenic enzyme controlled by SREBP-1 and energy status.
Sone H, Shimano H, Sakakura Y, Inoue N, Amemiya-Kudo M, Yahagi N, Osawa M, Suzuki H, Yokoo T, Takahashi A, Iida K, Toyoshima H, Iwama A, Yamada N. Sone H, et al. Am J Physiol Endocrinol Metab. 2002 Jan;282(1):E222-30. doi: 10.1152/ajpendo.00189.2001. Am J Physiol Endocrinol Metab. 2002. PMID: 11739104 - Hepatitis B virus-X protein upregulates the expression of p21waf1/cip1 and prolongs G1-->S transition via a p53-independent pathway in human hepatoma cells.
Park US, Park SK, Lee YI, Park JG, Lee YI. Park US, et al. Oncogene. 2000 Jul 13;19(30):3384-94. doi: 10.1038/sj.onc.1203674. Oncogene. 2000. PMID: 10918595 - Sterol regulatory element-binding protein family as global regulators of lipid synthetic genes in energy metabolism.
Shimano H. Shimano H. Vitam Horm. 2002;65:167-94. doi: 10.1016/s0083-6729(02)65064-2. Vitam Horm. 2002. PMID: 12481547 Review.
Cited by
- Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit.
Takeuchi Y, Yahagi N, Izumida Y, Nishi M, Kubota M, Teraoka Y, Yamamoto T, Matsuzaka T, Nakagawa Y, Sekiya M, Iizuka Y, Ohashi K, Osuga J, Gotoda T, Ishibashi S, Itaka K, Kataoka K, Nagai R, Yamada N, Kadowaki T, Shimano H. Takeuchi Y, et al. J Biol Chem. 2010 Apr 9;285(15):11681-91. doi: 10.1074/jbc.M109.096107. Epub 2010 Feb 9. J Biol Chem. 2010. PMID: 20145241 Free PMC article. - Sterol regulatory element-binding protein (SREBP)-1-mediated lipogenesis is involved in cell senescence.
Kim YM, Shin HT, Seo YH, Byun HO, Yoon SH, Lee IK, Hyun DH, Chung HY, Yoon G. Kim YM, et al. J Biol Chem. 2010 Sep 17;285(38):29069-77. doi: 10.1074/jbc.M110.120386. Epub 2010 Jul 8. J Biol Chem. 2010. PMID: 20615871 Free PMC article. - Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7α (Fbw7α) causes hepatosteatosis through Krüppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor γ2 (PPARγ2) pathway but not SREBP-1c protein in mice.
Kumadaki S, Karasawa T, Matsuzaka T, Ema M, Nakagawa Y, Nakakuki M, Saito R, Yahagi N, Iwasaki H, Sone H, Takekoshi K, Yatoh S, Kobayashi K, Takahashi A, Suzuki H, Takahashi S, Yamada N, Shimano H. Kumadaki S, et al. J Biol Chem. 2011 Nov 25;286(47):40835-46. doi: 10.1074/jbc.M111.235283. Epub 2011 Sep 12. J Biol Chem. 2011. PMID: 21911492 Free PMC article. - Proto-oncogene FBI-1 (Pokemon) and SREBP-1 synergistically activate transcription of fatty-acid synthase gene (FASN).
Choi WI, Jeon BN, Park H, Yoo JY, Kim YS, Koh DI, Kim MH, Kim YR, Lee CE, Kim KS, Osborne TF, Hur MW. Choi WI, et al. J Biol Chem. 2008 Oct 24;283(43):29341-54. doi: 10.1074/jbc.M802477200. Epub 2008 Aug 5. J Biol Chem. 2008. PMID: 18682402 Free PMC article. - p53 regulates the mevalonate pathway in human glioblastoma multiforme.
Laezza C, D'Alessandro A, Di Croce L, Picardi P, Ciaglia E, Pisanti S, Malfitano AM, Comegna M, Faraonio R, Gazzerro P, Bifulco M. Laezza C, et al. Cell Death Dis. 2015 Oct 15;6(10):e1909. doi: 10.1038/cddis.2015.279. Cell Death Dis. 2015. PMID: 26469958 Free PMC article.
References
- Amemiya-Kudo, M., H. Shimano, A. H. Hasty, N. Yahagi, T. Yoshikawa, T. Matsuzaka, H. Okazaki, Y. Tamura, Y. Iizuka, K. Ohashi, J. Osuga, K. Harada, T. Gotoda, R. Sato, S. Kimura, S. Ishibashi, and N. Yamada. 2002. Transcriptional activities of nuclear SREBP-1a, SREBP-1c, and SREBP-2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 43:1220-1235. - PubMed
- Amemiya-Kudo, M., H. Shimano, T. Yoshikawa, N. Yahagi, A. H. Hasty, H. Okazaki, Y. Tamura, F. Shionoiri, Y. Iizuka, K. Ohashi, J. Osuga, K. Harada, T. Gotoda, R. Sato, S. Kimura, S. Ishibashi, and N. Yamada. 2000. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 275:31078-31085. - PubMed
- Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. - PubMed
- Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases