Evolutionary constraints in conserved nongenic sequences of mammals - PubMed (original) (raw)
Evolutionary constraints in conserved nongenic sequences of mammals
Peter D Keightley et al. Genome Res. 2005 Oct.
Abstract
Mammalian genomes contain many highly conserved nongenic sequences (CNGs) whose functional significance is poorly understood. Sets of CNGs have previously been identified by selecting the most conserved elements from a chromosome or genome, but in these highly selected samples, conservation may be unrelated to purifying selection. Furthermore, conservation of CNGs may be caused by mutation rate variation rather than selective constraints. To account for the effect of selective sampling, we have examined conservation of CNGs in taxa whose evolution is largely independent of the taxa from which the CNGs were initially identified, and we have controlled for mutation rate variation in the genome. We show that selective constraints in CNGs and their flanks are about one-half as strong in hominids as in murids, implying that hominids have accumulated many slightly deleterious mutations in functionally important nongenic regions. This is likely to be a consequence of the low effective population size of hominids leading to a reduced effectiveness of selection. We estimate that there are one and two times as many conserved nucleotides in CNGs as in known protein-coding genes of hominids and murids, respectively. Polymorphism frequencies in CNGs indicate that purifying selection operates in these sequences. During hominid evolution, we estimate that a total of about three deleterious mutations in CNGs and protein-coding genes have been selectively eliminated per diploid genome each generation, implying that deleterious mutations are eliminated from populations non-independently and that sex is necessary for long-term population persistence.
Figures
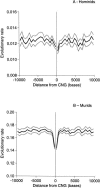
Figure 1.
Evolutionary rate calculated by the Tamura-Nei method in sequences flanking CNGs in (A) hominids and (B) murids. 95% confidence limits are shown in gray.
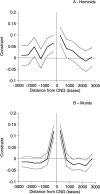
Figure 2.
Constraint in sequences flanking CNGs in (A) hominids and (B) murids. 95% confidence limits are show in gray.
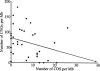
Figure 3.
Relationship between CNG density and coding sequence (CDS) density on the long arm of Chromosome 21, and the linear regression line.
Similar articles
- Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment.
Dermitzakis ET, Kirkness E, Schwarz S, Birney E, Reymond A, Antonarakis SE. Dermitzakis ET, et al. Genome Res. 2004 May;14(5):852-9. doi: 10.1101/gr.1934904. Epub 2004 Apr 12. Genome Res. 2004. PMID: 15078857 Free PMC article. - Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs).
Dermitzakis ET, Reymond A, Scamuffa N, Ucla C, Kirkness E, Rossier C, Antonarakis SE. Dermitzakis ET, et al. Science. 2003 Nov 7;302(5647):1033-5. doi: 10.1126/science.1087047. Epub 2003 Oct 2. Science. 2003. PMID: 14526086 - Evidence for widespread degradation of gene control regions in hominid genomes.
Keightley PD, Lercher MJ, Eyre-Walker A. Keightley PD, et al. PLoS Biol. 2005 Feb;3(2):e42. doi: 10.1371/journal.pbio.0030042. Epub 2005 Jan 25. PLoS Biol. 2005. PMID: 15678168 Free PMC article. - Nonadaptive processes in primate and human evolution.
Harris EE. Harris EE. Am J Phys Anthropol. 2010;143 Suppl 51:13-45. doi: 10.1002/ajpa.21439. Am J Phys Anthropol. 2010. PMID: 21086525 Review. - Raising the estimate of functional human sequences.
Pheasant M, Mattick JS. Pheasant M, et al. Genome Res. 2007 Sep;17(9):1245-53. doi: 10.1101/gr.6406307. Epub 2007 Aug 9. Genome Res. 2007. PMID: 17690206 Review.
Cited by
- Adaptive evolution of conserved noncoding elements in mammals.
Kim SY, Pritchard JK. Kim SY, et al. PLoS Genet. 2007 Sep;3(9):1572-86. doi: 10.1371/journal.pgen.0030147. Epub 2007 Jul 18. PLoS Genet. 2007. PMID: 17845075 Free PMC article. - Identification and characterization of new long conserved noncoding sequences in vertebrates.
Sakuraba Y, Kimura T, Masuya H, Noguchi H, Sezutsu H, Takahasi KR, Toyoda A, Fukumura R, Murata T, Sakaki Y, Yamamura M, Wakana S, Noda T, Shiroishi T, Gondo Y. Sakuraba Y, et al. Mamm Genome. 2008 Oct-Dec;19(10-12):703-12. doi: 10.1007/s00335-008-9152-7. Epub 2008 Nov 18. Mamm Genome. 2008. PMID: 19015917 - Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence.
Torgerson DG, Boyko AR, Hernandez RD, Indap A, Hu X, White TJ, Sninsky JJ, Cargill M, Adams MD, Bustamante CD, Clark AG. Torgerson DG, et al. PLoS Genet. 2009 Aug;5(8):e1000592. doi: 10.1371/journal.pgen.1000592. Epub 2009 Aug 7. PLoS Genet. 2009. PMID: 19662163 Free PMC article. - The genetics of inbreeding depression.
Charlesworth D, Willis JH. Charlesworth D, et al. Nat Rev Genet. 2009 Nov;10(11):783-96. doi: 10.1038/nrg2664. Nat Rev Genet. 2009. PMID: 19834483 Review. - The strength of selection on ultraconserved elements in the human genome.
Chen CT, Wang JC, Cohen BA. Chen CT, et al. Am J Hum Genet. 2007 Apr;80(4):692-704. doi: 10.1086/513149. Epub 2007 Feb 20. Am J Hum Genet. 2007. PMID: 17357075 Free PMC article.
References
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S. and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321-1325. - PubMed
- Casane, D., Boissinot, S., Chang, B.H.J., Shimmin, L.C., and Li, W.-H. 1997. Mutation pattern variation among regions of the primate genome. J. Mol. Evol. 45: 216-226. - PubMed
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420: 578-582. - PubMed
- Dermitzakis, E.T., Reymond, A., Scamuffa, N., Ucla, C., Kirkness, E., Rossier, C., and Antonarakis, S.E. 2003. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs). Science 302: 1033-1035. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources