De novo kinetochore assembly requires the centromeric histone H3 variant - PubMed (original) (raw)
De novo kinetochore assembly requires the centromeric histone H3 variant
Kimberly A Collins et al. Mol Biol Cell. 2005 Dec.
Abstract
Kinetochores mediate chromosome attachment to the mitotic spindle to ensure accurate chromosome segregation. Budding yeast is an excellent organism for kinetochore assembly studies because it has a simple defined centromere sequence responsible for the localization of >65 proteins. In addition, yeast is the only organism where a conditional centromere is available to allow studies of de novo kinetochore assembly. Using a conditional centromere, we found that yeast kinetochore assembly is not temporally restricted and can occur in both G1 phase and prometaphase. We performed the first investigation of kinetochore assembly in the absence of the centromeric histone H3 variant Cse4 and found that all proteins tested depend on Cse4 to localize. Consistent with this observation, Cse4-depleted cells had severe chromosome segregation defects. We therefore propose that yeast kinetochore assembly requires both centromeric DNA specificity and centromeric chromatin.
Figures
Figure 1.
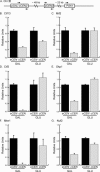
Kinetochore proteins from each subcomplex can assemble in G1. (A). Schematic of chromosome III (Chr III) indicates the position of the cCEN (pGAL-cCEN3) and the negative control locus (PGK1) with respect to the eCEN (eCEN3). Arrows indicate the PCR primer sets used to detect these loci in ChIP experiments shown in the subsequent figures. (B–G). Strains expressing Ctf13-myc13, Mif2-myc13, Ctf3-myc13, Okp1-myc13, Mtw1-myc13, and Nuf2-myc13 were arrested in G1 with αF and then transferred to media containing GAL or GLU. ChIP was performed with anti-myc antibody. Relative units indicate the eCEN:PGK1 ratio or the cCEN:PGK1 ratio and are normalized to an eCEN:PGK1 value of 1.0 for each experiment. All kinetochore proteins tested localize to the active cCEN in G1.
Figure 2.
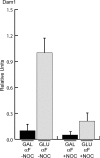
The G1 cCEN requires microtubules for kinetochore assembly. Cells expressing Dam1-myc9 were arrested in G1 with αFin galactose and then treated with (+NOC) or without (–NOC) the microtubule-depolymerizing drug nocodazole. Subsequently, the cultures were split into media containing GAL or GLU while maintaining the nocodazole treatment in αF. ChIP was performed with anti-myc antibody. Relative units indicate the cCEN:PGK1 ratio and the amount of Dam1-myc9 in GLU-NOC is normalized to a value of 1.0. Dam1 localization to the cCEN in G1 depends on microtubules.
Figure 3.
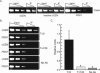
Cse4 localizes to the inactive cCEN and can be depleted using a Degron-Cse4 protein. (A) Degron-CSE4 pGAL-UBR1-myc cells were grown in galactose and arrested with nocodazole. ChIP was performed with anti-Cse4 antibody. Serial dilutions of lysates (input) and immunoprecipitated DNA (IP) are shown. Cse4 localizes to the inactive cCEN at levels similar to the eCEN. (B) Cells were grown as described above (A), and dox was added to repress transcription of _Degron_-CSE4. ChIP was performed using either anti-Cse4 antibody or no antibody at the indicated times after dox addition. Cse4 is depleted from the cCEN within 120 min. (C). Quantification of B. Relative units indicate the cCEN: PGK1 ratio.
Figure 4.
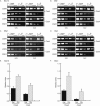
Cse4 is required for the full occupancy of inner and some central kinetochore components. (A–F). pGAL-UBR1-myc cCEN cells expressing the indicated myc-epitope tagged kinetochore protein and either wild-type CSE4 (+Cse4) or Degron-CSE4 (–Cse4) were grown in media containing galactose and arrested with nocodazole. Dox was added to repress transcription of _Degron_-CSE4, and cells were transferred into media containing GAL (inactive cCEN) or GLU (active cCEN) and analyzed by ChIP with anti-myc antibody. (E and F) Quantification of A and C. Relative units indicate the cCEN:PGK1 ratio. The cCEN:PGK1 value in GLU +Cse4 is normalized to a value of 1.0. Cse4 is required for the complete occupancy of Ndc10-myc13, Mif2-myc13, Mtw1-myc13, and Okp1-myc13 (also see Supplemental Figure 1, B and C).
Figure 5.
Cse4 is required to localize some central and outer kinetochore components. (A and B). pGAL-UBR1-myc cCEN cells and the indicated myc-epitope tagged kinetochore protein with either wild-type CSE4 (+Cse4) or Degron-CSE4 (–Cse4) were grown in media containing galactose (cCEN inactive) and arrested in prometaphase with nocodazole. Dox was added to repress transcription of _Degron_-CSE4, and cells were transferred into media containing GAL (inactive cCEN) or GLU, active cCEN) and analyzed by ChIP with anti-myc antibody. Cse4 is required to localize Ctf19-myc13 and Ndc80-myc13 to the cCEN. (C and D). Cse4 was depleted from the cCEN in cells expressing Dam1-myc9 or Ask1-myc13 by incubation with dox for 4 h. Cells were transferred to media containing GAL (inactive cCEN) or GLU (active cCEN) in the presence of dox and then harvested for ChIP with anti-myc antibody. Cse4 is required to localize Dam1-myc9 and Ask1-myc13. (E and F). Quantification of C and D. Relative units indicate the cCEN:PGK1 ratio. The cCEN:PGK1 value in GLU +Cse4 is normalized to a value of 1.0.
Figure 6.
Cse4 can be depleted from the eCEN. Cells expressing Degron-CSE4 (_Degron-CSE4 pGAL-UBR1-myc cse4_Δ _mad2_Δ) were arrested in αF in media containing galactose. Dox was added to repress transcription of Degron-CSE4, and then cells were released into the cell cycle in the absence of Cse4. Cells were harvested for Cse4 ChIP (A) and FACS (B) at –120 min (when dox was added) and 0, 30, 60, and 90 min after αF release. Cse4 is completely depleted from the eCEN within 90 min.
Figure 7.
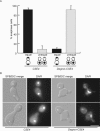
Cse4-depleted cells are defective in chromosome segregation. Cells expressing Degron-CSE4 (_mad2_Δ Spc42-GFP _cse4_Δ Degron-CSE4 pGAL-UBR1-myc) or wild-type Cse4 (_mad2_Δ Spc42-GFP pGAL-UBR1-myc) were grown in galactose and arrested in αF. Dox was added, and cells were released into the cell cycle in the absence of Cse4. After 150 min, cells were fixed and stained with DAPI. (A). At 150 min, cells that had segregated spindle pole bodies (anaphase cells) were scored as having equal DAPI masses (equal) or unequal DAPI masses (unequal). (B). Representative cells at the 150-min time point. The top images of the Degron-CSE4 show a cell with one, decondensed DAPI mass, and the bottom images represent a cell that has undergone unequal segregation where a small amount of DNA is in the bud. Bar, 5 μm.
Similar articles
- Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae.
Choy JS, Mishra PK, Au WC, Basrai MA. Choy JS, et al. Biochim Biophys Acta. 2012 Jul;1819(7):776-83. doi: 10.1016/j.bbagrm.2012.02.008. Epub 2012 Feb 16. Biochim Biophys Acta. 2012. PMID: 22366340 Free PMC article. Review. - Histone H3 localizes to the centromeric DNA in budding yeast.
Lochmann B, Ivanov D. Lochmann B, et al. PLoS Genet. 2012 May;8(5):e1002739. doi: 10.1371/journal.pgen.1002739. Epub 2012 May 31. PLoS Genet. 2012. PMID: 22693454 Free PMC article. - The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation.
Hsu JM, Huang J, Meluh PB, Laurent BC. Hsu JM, et al. Mol Cell Biol. 2003 May;23(9):3202-15. doi: 10.1128/MCB.23.9.3202-3215.2003. Mol Cell Biol. 2003. PMID: 12697820 Free PMC article. - Kinetochore function and chromosome segregation rely on critical residues in histones H3 and H4 in budding yeast.
Ng TM, Lenstra TL, Duggan N, Jiang S, Ceto S, Holstege FC, Dai J, Boeke JD, Biggins S. Ng TM, et al. Genetics. 2013 Nov;195(3):795-807. doi: 10.1534/genetics.113.152082. Epub 2013 Sep 13. Genetics. 2013. PMID: 24037263 Free PMC article. - Epigenetic regulation of centromeric chromatin: old dogs, new tricks?
Allshire RC, Karpen GH. Allshire RC, et al. Nat Rev Genet. 2008 Dec;9(12):923-37. doi: 10.1038/nrg2466. Nat Rev Genet. 2008. PMID: 19002142 Free PMC article. Review.
Cited by
- The overexpression of a Saccharomyces cerevisiae centromeric histone H3 variant mutant protein leads to a defect in kinetochore biorientation.
Collins KA, Camahort R, Seidel C, Gerton JL, Biggins S. Collins KA, et al. Genetics. 2007 Feb;175(2):513-25. doi: 10.1534/genetics.106.064410. Epub 2006 Dec 6. Genetics. 2007. PMID: 17151247 Free PMC article. - Synergistic Control of Kinetochore Protein Levels by Psh1 and Ubr2.
Herrero E, Thorpe PH. Herrero E, et al. PLoS Genet. 2016 Feb 18;12(2):e1005855. doi: 10.1371/journal.pgen.1005855. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26891228 Free PMC article. - Boolean gene regulatory network model of centromere function in Saccharomyces cerevisiae.
Haliki E, Alpagut Keskin N, Masalci O. Haliki E, et al. J Biol Phys. 2019 Sep;45(3):235-251. doi: 10.1007/s10867-019-09526-4. Epub 2019 Jun 7. J Biol Phys. 2019. PMID: 31175490 Free PMC article. - Control of the spindle checkpoint by lateral kinetochore attachment and limited Mad1 recruitment.
Krefman NI, Drubin DG, Barnes G. Krefman NI, et al. Mol Biol Cell. 2015 Jul 15;26(14):2620-39. doi: 10.1091/mbc.E15-05-0276. Epub 2015 May 28. Mol Biol Cell. 2015. PMID: 26023090 Free PMC article. - CaMtw1, a member of the evolutionarily conserved Mis12 kinetochore protein family, is required for efficient inner kinetochore assembly in the pathogenic yeast Candida albicans.
Roy B, Burrack LS, Lone MA, Berman J, Sanyal K. Roy B, et al. Mol Microbiol. 2011 Apr;80(1):14-32. doi: 10.1111/j.1365-2958.2011.07558.x. Epub 2011 Feb 10. Mol Microbiol. 2011. PMID: 21276093 Free PMC article.
References
- Biggins, S., and Walczak, C. E. (2003). Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13, R449–R460. - PubMed
- Buchwitz, B. J., Ahmad, K., Moore, L. L., Roth, M. B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547–548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases