Non-classical protein secretion in bacteria - PubMed (original) (raw)
Comparative Study
Non-classical protein secretion in bacteria
Jannick D Bendtsen et al. BMC Microbiol. 2005.
Abstract
Background: We present an overview of bacterial non-classical secretion and a prediction method for identification of proteins following signal peptide independent secretion pathways. We have compiled a list of proteins found extracellularly despite the absence of a signal peptide. Some of these proteins also have known roles in the cytoplasm, which means they could be so-called "moon-lightning" proteins having more than one function.
Results: A thorough literature search was conducted to compile a list of currently known bacterial non-classically secreted proteins. Pattern finding methods were applied to the sequences in order to identify putative signal sequences or motifs responsible for their secretion. We have found no signal or motif characteristic to any majority of the proteins in the compiled list of non-classically secreted proteins, and conclude that these proteins, indeed, seem to be secreted in a novel fashion. However, we also show that the apparently non-classically secreted proteins are still distinguished from cellular proteins by properties such as amino acid composition, secondary structure and disordered regions. Specifically, prediction of disorder reveals that bacterial secretory proteins are more structurally disordered than their cytoplasmic counterparts. Finally, artificial neural networks were used to construct protein feature based methods for identification of non-classically secreted proteins in both Gram-positive and Gram-negative bacteria.
Conclusion: We present a publicly available prediction method capable of discriminating between this group of proteins and other proteins, thus allowing for the identification of novel non-classically secreted proteins. We suggest candidates for non-classically secreted proteins in Escherichia coli and Bacillus subtilis. The prediction method is available online.
Figures
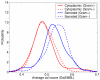
Figure 1
Bacterial secreted proteins are more disordered in structure than cytoplasmic proteins. The predicted number of coils (average per residue) by DisEMBL is higher for secreted proteins than for cytoplasmic ones. The tendency also holds for the two other measures of disorder predicted by DisEMBL (not shown).
Figure 2
Prediction on different proteins in different organisms. Four proteins from five bacterial species. Scores above 0.5 indicate predicted secretion of that particular protein. For details, please refer to the text.
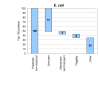
Figure 3
Putative non-classical secretory proteins. The top 100 scoring proteins are grouped based on annotation. Several groups have obvious relations to extracellular functions. For proteins with no annotation (grouped in 'Unknown'), our prediction method suggests an extracellular role. Proteins grouped in 'Other' may have an extracellular function, although this is not apparent in the current annotation.
Similar articles
- SecretP: identifying bacterial secreted proteins by fusing new features into Chou's pseudo-amino acid composition.
Yu L, Guo Y, Li Y, Li G, Li M, Luo J, Xiong W, Qin W. Yu L, et al. J Theor Biol. 2010 Nov 7;267(1):1-6. doi: 10.1016/j.jtbi.2010.08.001. Epub 2010 Aug 5. J Theor Biol. 2010. PMID: 20691704 - Feature-based reappraisal of the Bacillus subtilis exoproteome.
Tjalsma H. Tjalsma H. Proteomics. 2007 Jan;7(1):73-81. doi: 10.1002/pmic.200600520. Proteomics. 2007. PMID: 17149778 - Proteomics-based consensus prediction of protein retention in a bacterial membrane.
Tjalsma H, van Dijl JM. Tjalsma H, et al. Proteomics. 2005 Nov;5(17):4472-82. doi: 10.1002/pmic.200402080. Proteomics. 2005. PMID: 16220534 - Regulation of fatty acid metabolism in bacteria.
Fujita Y, Matsuoka H, Hirooka K. Fujita Y, et al. Mol Microbiol. 2007 Nov;66(4):829-39. doi: 10.1111/j.1365-2958.2007.05947.x. Epub 2007 Oct 2. Mol Microbiol. 2007. PMID: 17919287 Review. - Principle and potential applications of the non-classical protein secretory pathway in bacteria.
Kang Q, Zhang D. Kang Q, et al. Appl Microbiol Biotechnol. 2020 Feb;104(3):953-965. doi: 10.1007/s00253-019-10285-4. Epub 2019 Dec 18. Appl Microbiol Biotechnol. 2020. PMID: 31853566 Review.
Cited by
- Genomic comparison between Mycobacterium bovis and Mycobacterium microti and in silico analysis of peptide-based biomarkers for serodiagnosis.
Moens C, Bogaerts B, Lorente-Leal V, Vanneste K, De Keersmaecker SCJ, Roosens NHC, Mostin L, Fretin D, Marché S. Moens C, et al. Front Vet Sci. 2024 Sep 20;11:1446930. doi: 10.3389/fvets.2024.1446930. eCollection 2024. Front Vet Sci. 2024. PMID: 39372902 Free PMC article. - Aerobic adaptation and metabolic dynamics of Propionibacterium freudenreichii DSM 20271: insights from comparative transcriptomics and surfaceome analysis.
Loivamaa I, Sillanpää A, Deptula P, Chamlagain B, Edelmann M, Auvinen P, Nyman TA, Savijoki K, Piironen V, Varmanen P. Loivamaa I, et al. mSystems. 2024 Oct 22;9(10):e0061524. doi: 10.1128/msystems.00615-24. Epub 2024 Sep 30. mSystems. 2024. PMID: 39345151 Free PMC article. - Definition of the effector landscape across 13 phytoplasma proteomes with LEAPH and EffectorComb.
Calia G, Cestaro A, Schuler H, Janik K, Donati C, Moser M, Bottini S. Calia G, et al. NAR Genom Bioinform. 2024 Jul 30;6(3):lqae087. doi: 10.1093/nargab/lqae087. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39081684 Free PMC article. - APEX2-based proximity proteomic analysis identifies candidate interactors for Plasmodium falciparum knob-associated histidine-rich protein in infected erythrocytes.
Charneau S, de Oliveira LS, Zenonos Z, Hopp CS, Bastos IMD, Loew D, Lombard B, Pandolfo Silveira A, de Carvalho Nardeli Basílio Lobo G, Bao SN, Grellier P, Rayner JC. Charneau S, et al. Sci Rep. 2024 May 16;14(1):11242. doi: 10.1038/s41598-024-61295-w. Sci Rep. 2024. PMID: 38755230 Free PMC article. - The impact of Elaeagnus angustifolia root exudates on Parafrankia soli NRRL B-16219 exoproteome.
Kammoun I, Miotello G, Ben Slama K, Armengaud J, Ghodhbane-Gtari F, Gtari M. Kammoun I, et al. J Genomics. 2024 May 11;12:58-70. doi: 10.7150/jgen.93243. eCollection 2024. J Genomics. 2024. PMID: 38751381 Free PMC article.
References
- Rubartelli A, Bajetto A, Allavena G, Wollman E, Sitia R. Secretion of thioredoxin by normal and neoplastic cells through a leaderless secretory pathway. J Biol Chem. 1992;267:24161–24164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources