The geographic spread of the CCR5 Delta32 HIV-resistance allele - PubMed (original) (raw)
The geographic spread of the CCR5 Delta32 HIV-resistance allele
John Novembre et al. PLoS Biol. 2005 Nov.
Abstract
The Delta32 mutation at the CCR5 locus is a well-studied example of natural selection acting in humans. The mutation is found principally in Europe and western Asia, with higher frequencies generally in the north. Homozygous carriers of the Delta32 mutation are resistant to HIV-1 infection because the mutation prevents functional expression of the CCR5 chemokine receptor normally used by HIV-1 to enter CD4+ T cells. HIV has emerged only recently, but population genetic data strongly suggest Delta32 has been under intense selection for much of its evolutionary history. To understand how selection and dispersal have interacted during the history of the Delta32 allele, we implemented a spatially explicit model of the spread of Delta32. The model includes the effects of sampling, which we show can give rise to local peaks in observed allele frequencies. In addition, we show that with modest gradients in selection intensity, the origin of the Delta32 allele may be relatively far from the current areas of highest allele frequency. The geographic distribution of the Delta32 allele is consistent with previous reports of a strong selective advantage (>10%) for Delta32 carriers and of dispersal over relatively long distances (>100 km/generation). When selection is assumed to be uniform across Europe and western Asia, we find support for a northern European origin and long-range dispersal consistent with the Viking-mediated dispersal of Delta32 proposed by G. Lucotte and G. Mercier. However, when we allow for gradients in selection intensity, we estimate the origin to be outside of northern Europe and selection intensities to be strongest in the northwest. Our results describe the evolutionary history of the Delta32 allele and establish a general methodology for studying the geographic distribution of selected alleles.
Figures
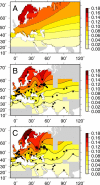
Figure 1. Shaded Contour Map of Δ32 Allele Frequency Data
The sampling locations are marked by black points. The interpolation is masked in regions where data are unavailable.
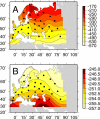
Figure 2. An Example of the Allele Frequency Surface and Simulated Data
(A) The underlying allele frequency surface generated by the PDE model using MLEs for the parameters. The coarseness of the surface and irregular coastlines are due to the resolution of the simulated habitat (see Figure S1). (B and C) Two replicates of simulated data obtained using the same sampling locales and sample sizes as in the dataset and displayed using the same interpolation methods and contours as in Figure 1. The results show that underlying smooth, unimodal allele frequency surfaces can give rise to irregular, multimodal observed allele frequency surfaces.
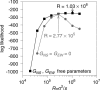
Figure 3. Profile Likelihood for R
The grey line shows the log profile likelihood for R when selection is assumed to be uniform spatially (GNS = GEW = 0). The MLE of R in this case is 2.77 × 105 with a log-likelihood of −263.0. The black line shows the profile likelihood when selection gradients are incorporated into the model (GNS and GEW are free parameters). The corresponding MLE of R is 1.03 × 106 with a log likelihood of −247.7.
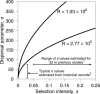
Figure 4. The Dispersal Parameter σ as a Function of the Selection Intensity s
The curves are drawn for the two MLEs of R and labeled accordingly. 1Based on estimates in [2] and [3]. 2From Table 1 of [27].
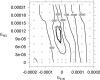
Figure 5. Profile Likelihood Surface for GNS and GEW
The plus signs indicate locations where the likelihood was evaluated. The dark contour at −250 marks the −2 log-likelihood support region for the estimates of GNS and GEW.
Figure 6. Likelihood Surfaces for the Origin of the Allele
(A) Assuming selection intensity is uniform spatially (i.e., GNS = GEW = 0). (B) Allowing for north–south and east–west spatial gradients in selection (i.e., GNS and GEW are free parameters). Likelihoods were calculated at each of the black points and the surface was obtained by interpolation.
Similar articles
- The evolutionary history of the CCR5-Delta32 HIV-resistance mutation.
Galvani AP, Novembre J. Galvani AP, et al. Microbes Infect. 2005 Feb;7(2):302-9. doi: 10.1016/j.micinf.2004.12.006. Epub 2005 Jan 8. Microbes Infect. 2005. PMID: 15715976 Review. - More about the Viking hypothesis of origin of the delta32 mutation in the CCR5 gene conferring resistance to HIV-1 infection.
Lucotte G, Dieterlen F. Lucotte G, et al. Infect Genet Evol. 2003 Nov;3(4):293-5. doi: 10.1016/j.meegid.2003.07.001. Infect Genet Evol. 2003. PMID: 14636691 - Is the European spatial distribution of the HIV-1-resistant CCR5-Delta32 allele formed by a breakdown of the pathocenosis due to the historical Roman expansion?
Faure E, Royer-Carenzi M. Faure E, et al. Infect Genet Evol. 2008 Dec;8(6):864-74. doi: 10.1016/j.meegid.2008.08.007. Epub 2008 Aug 27. Infect Genet Evol. 2008. PMID: 18790087 - The case for selection at CCR5-Delta32.
Sabeti PC, Walsh E, Schaffner SF, Varilly P, Fry B, Hutcheson HB, Cullen M, Mikkelsen TS, Roy J, Patterson N, Cooper R, Reich D, Altshuler D, O'Brien S, Lander ES. Sabeti PC, et al. PLoS Biol. 2005 Nov;3(11):e378. doi: 10.1371/journal.pbio.0030378. Epub 2005 Nov 1. PLoS Biol. 2005. PMID: 16248677 Free PMC article. - HIV-1 Infection in Persons Homozygous for CCR5-Δ32 Allele: The Next Case and the Review.
Smoleń-Dzirba J, Rosińska M, Janiec J, Beniowski M, Cycoń M, Bratosiewicz-Wąsik J, Wąsik TJ. Smoleń-Dzirba J, et al. AIDS Rev. 2017 Dec;19(4):219-230. AIDS Rev. 2017. PMID: 28534889 Review.
Cited by
- Unveiling ancestral threads: Exploring CCR5 ∆32 mutation frequencies in Colombian populations for HIV/AIDS therapeutics.
Barrios-Navas A, Nguyen TL, Gallo JE, Mariño-Ramírez L, Soto JMS, Sánchez A, Jordan IK, Valderrama-Aguirre A. Barrios-Navas A, et al. Infect Genet Evol. 2024 Nov;125:105680. doi: 10.1016/j.meegid.2024.105680. Epub 2024 Oct 5. Infect Genet Evol. 2024. PMID: 39374819 - Pervasive findings of directional selection realize the promise of ancient DNA to elucidate human adaptation.
Akbari A, Barton AR, Gazal S, Li Z, Kariminejad M, Perry A, Zeng Y, Mittnik A, Patterson N, Mah M, Zhou X, Price AL, Lander ES, Pinhasi R, Rohland N, Mallick S, Reich D. Akbari A, et al. bioRxiv [Preprint]. 2024 Sep 15:2024.09.14.613021. doi: 10.1101/2024.09.14.613021. bioRxiv. 2024. PMID: 39314480 Free PMC article. Preprint. - Building genomic capacity for precision health in Africa.
Olono A, Mitesser V, Happi A, Happi C. Olono A, et al. Nat Med. 2024 Jul;30(7):1856-1864. doi: 10.1038/s41591-024-03081-9. Epub 2024 Jul 3. Nat Med. 2024. PMID: 38961224 Review. - Gaining momentum: stem cell therapies for HIV cure.
Buck AM, LaFranchi BH, Henrich TJ. Buck AM, et al. Curr Opin HIV AIDS. 2024 Jul 1;19(4):194-200. doi: 10.1097/COH.0000000000000859. Epub 2024 Apr 26. Curr Opin HIV AIDS. 2024. PMID: 38686850 Free PMC article. Review. - Legacy of a magic gene-CCR5-∆32: From discovery to clinical benefit in a generation.
OBrien SJ. OBrien SJ. Proc Natl Acad Sci U S A. 2024 Mar 19;121(12):e2321907121. doi: 10.1073/pnas.2321907121. Epub 2024 Mar 8. Proc Natl Acad Sci U S A. 2024. PMID: 38457490 Free PMC article.
References
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. - PubMed
- Slatkin M. Simulating genealogies of selected alleles in a population of variable size. Genet Res. 2001;78:49–57. - PubMed
- Hummel S, Schmidt D, Kremeyer B, Herrmann B, Oppermann M. Detection of the CCR5-Delta32 HIV resistance gene in Bronze Age skeletons. Genes Immun. 2005;6:371–374. - PubMed
- Libert F, Cochaux P, Beckman G, Samson M, Aksenova M, et al. The delta CCR5 mutation conferring protection against HIV-1 in Caucasian populations has a single and recent origin in Northeastern Europe. Hum Mol Genet. 1998;7:399–406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials