Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution - PubMed (original) (raw)
Comparative Study
Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution
Xing Zhang et al. Structure. 2005 Oct.
Abstract
Reovirus is a useful model for addressing the molecular basis of membrane penetration by one of the larger nonenveloped animal viruses. We now report the structure of the reovirus virion at approximately 7.0 A resolution as obtained by electron cryomicroscopy and three-dimensional image reconstruction. Several features of the myristoylated outer capsid protein mu1, not seen in a previous X-ray crystal structure of the mu1-sigma3 heterohexamer, are evident in the virion. These features appear to be important for stabilizing the outer capsid, regulating the conformational changes in mu1 that accompany perforation of target membranes, and contributing directly to membrane penetration during cell entry.
Figures
Figure 1
Sequence and Secondary Structures of Reovirus μ1 Protein Primary sequence of reovirus μ1 protein (GenBank AAM10735) is shown in single-letter code with position numbers above. Three segments of disordered residues in the X-ray crystal model of the μ1-σ3 heterohexamer (Liemann et al., 2002) are indicated by larger, bold letters and dashed lines. Secondary structures extracted from the crystal structure are noted above the other sequences (α helices, cylinders; β strands, arrows). An arrowhead marks the autocleavage site between residues 42 and 43. The μ1 protein is N-terminally myristoylated, presumably on Gly2 after removal of Met1 (Nibert et al., 1991b; Tillotson and Shatkin, 1992).
Figure 2
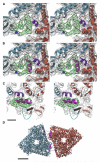
Cryo-EM Reconstruction of Reovirus Virion and Atomic Models of Virion and ISVP (A) Shaded surface representation of 7.0 Å cryo-EM reconstruction of reovirus virion, viewed along an icosahedral 2-fold axis. The 200 Å scale bar also applies to (B) and (C) and represents 100 Å for (D). (B) Atomic model of reovirus virion viewed as in (A). Proteins are differentiated by color: σ3 monomers, blue; λ2 pentamers, magenta; μ1.Q trimers, orange; μ1.R trimers, cyan; μ1.S trimers, pink; and μ1.T trimers, brown. The μ1 trimers are partially obscured by σ3 in the virion, but are clearly seen in the ISVP model in (C). P2 and P3 channels through the outer capsid are labeled; the former is bounded by four μ1 trimers and λ2, and the latter is bounded by six μ1 trimers (Dryden et al., 1993; Metcalf et al., 1991). One asymmetric unit, visible in this panel as well as in (C) and (D), is outlined with a white triangle. (C) Same as shown in (B), but minus the σ3 subunits in order to model the ISVP. The different types of μ1 trimers that surround the P2 and P3 channels indicated in (B) are now labeled with letters. The two S trimers related by the icosahedral 2-fold axis are labeled S1 and S2 to signify that they have distinct interactions with the underlying σ2 monomer (not visible). (D) Close-up view of a portion of the ISVP model shown in (C). Green symbols (pentagons, triangle, and oval) identify the icosahedral five-, three-, and 2-fold axes, respectively. (E) Close-up stereo view of part of a μ1.T trimer. Cryo-EM densities attributable to μ1.T in the 7.0 Å map of the virion (gray net, contoured at 2.0 σ) have been fitted with a model of the μ1 trimer (ribbon traces colored in purple, orange, and cyan for the three respective subunits in this trimer) derived from the 2.8 Å X-ray crystal structure (Liemann et al., 2002). Scale bar, 10 Å.
Figure 3
Resolutions of Virion and Averaged μ1 Subunit Maps Plots of the Fourier-shell correlation (FSC) as a function of resolution are depicted for the virion capsids (dashed curve), the μ1.T trimer densities (solid curve), and the averaged μ1 subunit densities (thick solid curve). Based on a conservative threshold criterion (solid horizontal line for FSC = 0.5 [Rosenthal and Henderson, 2003]), the effective resolutions of the virion capsids, μ1.T trimer, and averaged μ1 subunit are approximately 7.0, 6.9, and 6.7 Å, respectively. Based on a less stringent, noise-limited criterion (dashed horizontal line for FSC = 0.143 [Rosenthal and Henderson, 2003]), the effective resolutions are approximately 6.8, 6.7, and better than 6.0 Å, respectively. The FSC for the μ1 average exceeds that for the other two density maps at all resolutions, indicating that all features ≥ 6.0 Å are better represented in the average.
Figure 4
Cryo-EM Densities of μ1 before and after Averaging (A) Close-up stereo view of a portion of the μ1.T trimer before nonicosahedral averaging of the cryo-EM densities. The densities (gray net, contoured at 2.0 σ) have been fitted with the X-ray crystal model of the μ1 trimer (purple and cyan ribbon traces for two respective μ1 subunits from the same trimer). At this contour level, densities attributable to residues Pro316–Ala319 (red and yellow spheres, respectively, connected by purple trace) are discontinuous, and there is little density attributable to the side chain of Trp333 (ball and stick model). (B) Same as shown in (A), but for the nonicosahedral average of seven μ1 subunits (see Experimental Procedures). At a contour level of 2.0 σ, the cryo-EM densities attributable to residues Pro316–Ala319 are now nearly continuous, and there is somewhat more density attributable to the side chain of Trp333 (the latter being more evident when viewed from a different angle than the one chosen to highlight the 316–319 region in this figure). The 5 Å scale bar also applies to (A).
Figure 5
Cryo-EM Densities in the P3 and P2 Channels of Reovirus Virion (A–D) Cryo-EM densities (gray net) are contoured at 0.8 σ. Colors and labels correspond to those in Figures 2B–2D. Ribbon traces depict the fitted X-ray crystal models of the (A–D) μ1 trimers (Q, orange; R, cyan; S, pink, and T, brown), (B and D) associated σ3 monomers (blue), a (C and D) λ2 pentamer (magenta), and (B and D) σ2 monomers (purple). Pro675, the last residue visible in the μ1 crystal structure, is identified with a green ball for the μ1 subunits that project it into the highlighted (A and B) P3 and (C and D) P2 channels. “Hub-and-spokes” cryo-EM densities, devoid of fitted models but abutting Pro675, are visible in the channels, with six spokes in the P3 structure and four spokes in the P2 structure. (A) The slab shown in this panel is from the region bounded by dashed lines in (B). A radial channel through the P3 hub is more evident at lower resolutions (8–9 Å). (B) P3 channel, side view. The slab is from the region bounded by dashed lines in (A). (C) P2 channel, top view. The slab is from the region bounded by dashed lines in (D). (D) P2 channel, side view. The slab is from the region bounded by dashed lines in (C). The 25 Å scale bar applies to all panels.
Figure 6
Cryo-EM Densities for Residues 72–96 in the Averaged μ1 Cryo-EM Map (A) Close-up stereo view of a region near the base of two adjacent μ1 trimers, looking toward the virion interior. Cryo-EM densities (gray net, contoured at 2.0 σ) are fitted with the X-ray crystal models of two μ1 trimers (light-blue and brown ribbon traces, respectively). Extra cryo-EM densities forming a U-shaped loop (green net) were assigned to residues 72–96. Black and blue spheres, abutting the two ends of the U-shaped densities, identify the positions of residues Ile71 and Asp97, respectively, in the light-blue subunit. A short α helix (residues 122–127), colored purple in the otherwise brown subunit, is slightly displaced from the attributed cryo-EM densities, apparently owing to contacts with the 72–96 loop. (B) Same as shown in (A), but with a tentative ribbon trace (magenta), including an α helix, built into the extra cryo-EM densities ascribed to residues 72–96. (C) Same as shown in (B), but viewed in an orthogonal direction, from the left side of (B) to illustrate apparent interactions between the 72–96 (magenta) and the 51–62 (cyan) loops. The 51–62 loop is clearly displaced from the associated cryo-EM densities. A β strand formed by residues 63–65 (black) also contacts the 72–96 loop. The 10 Å scale bar also applies to (A) and (B). (D) Bottom view (from virion interior) of two adjacent μ1 trimers (light-blue and brown) in the atomic model. Putative traces for the two 72–96 loops at the trimer-trimer interface are shown in magenta. The scale bar is 25 Å.
Figure 7
Autocleavage Site in the Averaged μ1 Cryo-EM Map Portions of two μ1 subunits from the same trimer (indicated by purple and orange traces for the X-ray crystal models of these subunits) are shown in a close-up stereo view. The positions of Asn42, Pro43, and Gly45, as found in the cleaved μ1 subunits of crystallized μ1-σ3 heterohexamers (Liemann et al., 2002), are indicated with magenta, blue, and green spheres, respectively. Asn42 lies within the cryo-EM densities (gray net, contoured at 1.8 σ), whereas Pro43 and Gly44 (no sphere) lie outside, and these observations hold true over a broad range of contour levels (data not shown). Furthermore, unlike in the X-ray crystal structure, a continuous arch of density bridges residues 42–45 in the cryo-EM map, suggesting that the Asn42/Pro43 peptide bond remains uncleaved. The red trace within this arch models the putative path of μ1 residues 42–45 within the virion. The scale bar is 5 Å.
Figure 8
Cryo-EM Densities for N-Terminal Eight Residues of μ1 and Residues 580–586 of λ2 (A) Close-up stereoview of the cryoEM densities (gray net) and fitted X-ray crystal models (light-blue and brown ribbon traces) at the interface between two μ1 trimers. Extra cryo-EM densities (green net) have been assigned to the N-terminal eight residues of one μ1 subunit from the orange trimer. The position of Thr10 is marked by a blue sphere, residues Gly2 to Gln9 are modeled as a blue loop, and the N-terminal myristoyl group is represented as a space-filling model (red) occupying the β-octylglucoside pocket. Densities shown here, for the averaged μ1 subunits, are contoured at 1.4 σ. The 10 Å scale bar also applies to (C) and (D). (B) Same as shown in Figure 6D, but viewed in an orthogonal direction (from the bottom of Figure 6D) to show a full side view of the pair of μ1 trimers. Also highlighted are the putative trace for N-terminal residues 2–9 (blue) and the space-filling model for the N-terminal myristoyl group (red), for the one subunit in each trimer for which this region is positioned near the highlighted trimer-trimer interface. The scale bar is 25 Å. (C) Close-up view of the 7.0 Å virion reconstruction showing cryo-EM densities (gray net, contoured at 1.6 σ) and fitted X-ray models (ribbon traces) at the interface between the μ1.Q trimer (orange) and λ2 (magenta). Extra cryo-EM densities (green net) have been assigned to the 580–586 loop (cyan trace) of the λ2 methylase-1 domain and to approximately two residues of the N terminus of μ1 (blue sphere marks the position of Thr10). The short, purple-colored α helix in μ1 (residues 122–127) contacts the upper surface of the 580–586 loop of λ2. Conformation of the λ2 loop in the X-ray model of the core particle (Reinisch et al., 2000) places the loop outside the extra density, which indicates that interactions of λ2 with the neighboring μ1 subunit in virions results in a new loop conformation. (D) Same as shown in (C), but with a putative model (red trace) showing a proposed conformation of the 580–586 loop of λ2 fitted into the extra cryo-EM densities.
Figure 9
Contacts between μ1-σ3 Heterohexamers and between the μ1-σ3 Heterohexamer and λ2 Pentamer (A) Space-filling model of the μ1-σ3.S heterohexamer viewed from the direction of the surface that contacts a μ1-σ3.Q heterohexamer. Three σ3 subunits (blue) sit atop the intertwined red, green, and gray μ1 subunits in the S heterohexamer. White highlights those residues in μ1 and σ3 that contact μ1 subunits in the Q heterohexamer. Orange highlights those residues in σ3 that contact σ3 subunits in the Q heterohexamer. Magenta highlights those residues at the bottom of μ1 that contact σ2 subunits in the core (not shown). (B) Same as shown in (A), but for a view of the μ1-σ3.Q heterohexamer surface that contacts the μ1-σ3.S heterohexamer. The color scheme is identical to that in (A). (C) Same as shown in (B), but for a view of the μ1-σ3.Q heterohexamer surface that contacts the λ2 pentamer. White and purple highlight those residues in μ1 that contact λ2; white highlights residues that, on the other two faces of the Q heterohexamer, contact μ1 subunits in the R and S heterohexamers, whereas purple highlights residues that uniquely contact λ2. The remaining color scheme is the same as in (A). The 50 Å scale bar applies to all panels.
References
- Attoui H, Fang Q, Jaafar FM, Cantaloube JF, Biagini P, De Micco P, De Lamballerie X. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae) J. Gen. Virol. 2002;83:1941–1951. - PubMed
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. - PubMed
- Chacón P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 2002;317:375–384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM033050/GM/NIGMS NIH HHS/United States
- R37 CA013202/CA/NCI NIH HHS/United States
- P01 AI045976/AI/NIAID NIH HHS/United States
- R01 AI046440/AI/NIAID NIH HHS/United States
- R01 AI47904/AI/NIAID NIH HHS/United States
- R01 GM33050/GM/NIGMS NIH HHS/United States
- R01 AI46440/AI/NIAID NIH HHS/United States
- R01 CA013202/CA/NCI NIH HHS/United States
- R01 AI047904/AI/NIAID NIH HHS/United States
- R01 GM62580/GM/NIGMS NIH HHS/United States
- R01 CA13202/CA/NCI NIH HHS/United States
- R37 GM033050/GM/NIGMS NIH HHS/United States
- P01 GM062580/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases