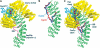Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling - PubMed (original) (raw)
Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling
Yoshiyuki Matsuura et al. EMBO J. 2005.
Abstract
Nuclear import of proteins containing classical nuclear localization signals (NLS) is mediated by the importin-alpha:beta complex that binds cargo in the cytoplasm and facilitates its passage through nuclear pores, after which nuclear RanGTP dissociates the import complex and the importins are recycled. In vertebrates, import is stimulated by nucleoporin Nup50, which has been proposed to accompany the import complex through nuclear pores. However, we show here that the Nup50 N-terminal domain actively displaces NLSs from importin-alpha, which would be more consistent with Nup50 functioning to coordinate import complex disassembly and importin recycling. The crystal structure of the importin-alpha:Nup50 complex shows that Nup50 binds at two sites on importin-alpha. One site overlaps the secondary NLS-binding site, whereas the second extends along the importin-alpha C-terminus. Mutagenesis indicates that interaction at both sites is required for Nup50 to displace NLSs. The Cse1p:Kap60p:RanGTP complex structure suggests how Nup50 is then displaced on formation of the importin-alpha export complex. These results provide a rationale for understanding the series of interactions that orchestrate the terminal steps of nuclear protein import.
Figures
Figure 1
Nup50 displaces classical NLSs from importin-α. (A) Solid phase binding assay with GST-Nup50 (residues 1–109) or NLS-GFP immobilized on the microtitre plate. Each data point was performed in duplicate and error bars represent s.e.m. NP stands for nucleoplasmin. (B) Pulldown competition assays. Immobilized GST-SV40 NLS (lanes 1–6) or GST-NP NLS (lanes 7–12) was incubated with 6.6 μM ΔIBB importin-α (lanes 1, 2, 7, and 8), or with 8 μM importins α and β (lanes 3, 4, 9, and 10), or with 37 μM importin-α alone (lanes 5, 6, 11, and 12), unbound material washed away and then treated with buffer alone (lanes 1, 3, 5, 7, 9, and 11) or with 20 μM Nup50 (residues 1–109) (lanes 2, 4, 6, 8, 10, and 12). (C) Equilibrium competition between Nup50 and the monopartite SV40 NLS in solution. Emission profiles of 0.2 μM BFP-ΔIBB importin-α and 0.2 μM SV40 NLS-GFP with (black) or without (red) 1 μM Nup50 (residues 1–109). (D) Equilibrium competition between Nup50 and the bipartite NP NLS in solution. Emission profiles of 0.2 μM BFP-ΔIBB importin-α and 0.2 μM NP NLS-GFP with (black) or without (red) 1 μM Nup50 (residues 1–109). (E) Stopped-flow traces following the dissociation kinetics of SV40 NLS from importin-α, 0.2 μM BFP-ΔIBB importin-α and 0.2 μM SV40 NLS-GFP alone (black points), or with 4 μM ΔIBB importin-α (red points), or with 1 μM Nup50 (residues 109) (blue points). Fitted exponential curves are superimposed. (F) Stopped-flow traces following the dissociation kinetics of NP NLS from importin-α, 0.2 μM BFP-ΔIBB importin-α and 0.2 μM NP NLS-GFP alone (black points), or with 4 μM ΔIBB importin-α (red points), or with 1 μM Nup50 (residues 109) (blue points). Fitted exponential curves are superimposed.
Figure 2
Crystal structure of the Nup50:importin-α complex and competition assays using structure-based mutants showed that Nup50 binds at two sites on importin-α, and the NLS displacement by Nup50 requires interaction with both sites on importin-α. (A) Overview of the Nup50:importin-α complex with the 2_F_o−_F_c map around Nup50 contoured at 1σ superimposed. Nup50 residues are shown in ball-and-stick format. (B) Electrostatic potential on importin-α, with Nup50 removed, shaded from −9 kT/e (red) to +9 kT/e (blue) calculated using GRASP (Nicholls et al, 1991). (C) FRET-based competition assay. Emission profiles of 0.2 μM BFP-ΔIBB importin-α and 0.2 μM SV40 NLS-GFP without (red) 1 μM Nup50 (residues 1–109) or with (black) 1 μM Nup50 (residues 1–109, wild type) or with (blue) 1 μM Nup50 (residues 1–109, K3E/R4D), or with (green) 1 μM Nup50 (residues 1–109, R38A/R45D). (D) FRET-based competition assay. Emission profiles of 0.2 μM BFP-ΔIBB importin-α and 0.2 μM NP NLS-GFP without (red) 1 μM Nup50 (residues 1–109) or with (black) 1 μM Nup50 (residues 1–109, wild type) or with (blue) 1 μM Nup50 (residues 1–109, K3E/R4D) or with (green) 1 μM Nup50 (residues 1–109, R38A/R45D).
Figure 3
Pulldown competition assay. Immobilized GST-SV40 NLS (lanes 1–3) or GST-NP NLS (lanes 4–6) was incubated with 6.6 μM ΔIBB importin-α, unbound protein washed away and then treated with 20 μM Nup50 (residues 1–109, wild type) (lanes 1 and 4) or with 20 μM Nup50 (residues 1–109, K3E/R4D) (lanes 2 and 5), or with 20 μM Nup50 (residues 1–109, R38A/R45D) (lanes 3 and 6).
Figure 4
Conserved interactions between Nup50 (Nup2p) and importin-α (Kap60p). (A) Revised model of the ΔIBB-Kap60p:Nup2p (residues 1–51) complex with the 2_F_o−_F_c map around Nup2p contoured at 1σ superimposed. Nup2p residues are shown in ball-and-stick format. (B) Superimposition of the main chains of Nup50 (magenta), Nup2p (green), SV40 NLS (cyan), and NP NLS (yellow) on the surface of ΔIBB importin-α. (C) Sequence alignment of yeast Nup2p and mouse Nup50 at the N-terminal importin-α-binding domain. (D) Mutants in the Nup2p residues analogous to those identified in the two importin-α-binding regions of Nup50 do not displace the monopartite or bipartite NLSs from yeast importin-α (Kap60p). Emission profiles of 0.2 μM BFP-ΔIBB-Kap60p and 0.2 μM SV40 NLS-GFP without (red) 2 μM Nup2p (residues 1–51) or with (black) 2 μM Nup2p (residues 1–51, wild type) or with (blue) 2 μM Nup2p (residues 1–51, K3E/R4D), or with (green) 2 μM Nup2p (residues 1–51, R38A/R39A/R46A/R47A). (E) Emission profiles of 0.2 μM BFP-ΔIBB-Kap60p and 0.2 μM NP NLS-GFP without (red) 2 μM Nup2p (residues 1–51) or with (black) 2 μM Nup2p (residues 1–51, wild type) or with (blue) 2 μM Nup2p (residues 1–51, K3E/R4D), or with (green) 2 μM Nup2p (residues 1–51, R38A/R39A/R46A/R47A).
Figure 5
Nup50 (Nup2p) binds to sites crucial for the formation of nuclear export complex of importin-α (Kap60p) with CAS (Cse1p) and RanGTP. (A) The Cse1p:Kap60p:RanGTP complex (Matsuura and Stewart, 2004). (B) Nup50 (red) and Nup2p (blue) are superimposed on the Kap60p ARM repeats in the Cse1p:Kap60p:RanGTP complex. (C) (A) and (B) superimposed. Cse1p, yellow; Ran, light blue; Kap60p IBB, pink; Kap60p ARM repeats, green. GTP is shown as space-filling model.
Figure 6
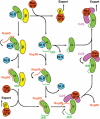
Schematic illustration of the possible ways in which NLS displacement and importin-α recycling could be mediated by Nup50, CAS and RanGTP. The disassembly of nuclear import complex and the assembly of nuclear export complexes could happen with or without Nup50. In the absence of Nup50, RanGTP would dissociate the IBB domain from importin-β, the NLS is released from importin-α by the IBB domain, and the cargo-free importins α and β are exported to cytoplasm separately. In the presence of Nup50, Nup50 could accelerate the NLS release and also act as a platform on which importin-α export complex is assembled. The active displacement of NLS by Nup50 probably occurs in two steps. First, Nup50 would make a transient complex with incoming importin-α–β:NLS complex by binding to the C-terminus of importin-α. The N-terminus of Nup50 would then displace NLS, and after importin-β is dissociated by RanGTP, the IBB domain, CAS and RanGTP cooperate to release importin-α from Nup50. Nup50 could also displace NLS from importin-α after the IBB domain is released from importin-β by RanGTP, and this may be important for dissociation of strongly bound bipartite cargoes for which the IBB domain alone may not be enough to achieve quick release. Thus, several parallel pathways are conceivable. These are not mutually exclusive and the presence of Nup50 at the nuclear face of the NPCs would increase the overall rate of nuclear trafficking, even though Nup50 may not be absolutely required for trafficking because of functional redundancy.
Similar articles
- The importin β binding domain as a master regulator of nucleocytoplasmic transport.
Lott K, Cingolani G. Lott K, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review. - Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import.
Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. Matsuura Y, et al. EMBO J. 2003 Oct 15;22(20):5358-69. doi: 10.1093/emboj/cdg538. EMBO J. 2003. PMID: 14532109 Free PMC article. - Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import.
Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG. Lindsay ME, et al. Cell. 2002 Aug 9;110(3):349-60. doi: 10.1016/s0092-8674(02)00836-x. Cell. 2002. PMID: 12176322 - Structural basis for the assembly of a nuclear export complex.
Matsuura Y, Stewart M. Matsuura Y, et al. Nature. 2004 Dec 16;432(7019):872-7. doi: 10.1038/nature03144. Nature. 2004. PMID: 15602554 - Npap60: a new player in nuclear protein import.
Moore MS. Moore MS. Trends Cell Biol. 2003 Feb;13(2):61-4. doi: 10.1016/s0962-8924(02)00044-2. Trends Cell Biol. 2003. PMID: 12559755 Review.
Cited by
- Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin α5.
Pumroy RA, Nardozzi JD, Hart DJ, Root MJ, Cingolani G. Pumroy RA, et al. J Biol Chem. 2012 Jan 13;287(3):2022-31. doi: 10.1074/jbc.M111.315838. Epub 2011 Nov 30. J Biol Chem. 2012. PMID: 22130666 Free PMC article. - The importin β binding domain as a master regulator of nucleocytoplasmic transport.
Lott K, Cingolani G. Lott K, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review. - PORE-ing over ERK substrates.
Ahn NG. Ahn NG. Nat Struct Mol Biol. 2009 Oct;16(10):1004-5. doi: 10.1038/nsmb1009-1004. Nat Struct Mol Biol. 2009. PMID: 19809488 No abstract available. - The nucleoporin Nup50 activates the Ran guanine nucleotide exchange factor RCC1 to promote NPC assembly at the end of mitosis.
Holzer G, De Magistris P, Gramminger C, Sachdev R, Magalska A, Schooley A, Scheufen A, Lennartz B, Tatarek-Nossol M, Lue H, Linder MI, Kutay U, Preisinger C, Moreno-Andres D, Antonin W. Holzer G, et al. EMBO J. 2021 Dec 1;40(23):e108788. doi: 10.15252/embj.2021108788. Epub 2021 Nov 2. EMBO J. 2021. PMID: 34725842 Free PMC article. - Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo.
Fox AM, Ciziene D, McLaughlin SH, Stewart M. Fox AM, et al. J Biol Chem. 2011 Aug 19;286(33):29325-29335. doi: 10.1074/jbc.M111.245092. Epub 2011 Jun 27. J Biol Chem. 2011. PMID: 21708948 Free PMC article.
References
- Bayliss R, Littlewood T, Stewart M (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 102: 99–108 - PubMed
- Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M (2002) GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J Biol Chem 277: 50597–50606 - PubMed
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
- Catimel B, Teh T, Fontes MR, Jennings IG, Jans DA, Howlett GJ, Nice EC, Kobe B (2001) Biophysical characterization of interactions involving importin-α during nuclear import. J Biol Chem 276: 34189–34198 - PubMed
- Cingolani G, Petosa C, Weis K, Muller CW (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature 399: 221–229 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases