The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles - PubMed (original) (raw)
The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles
Danso Ako-Adjei et al. J Virol. 2005 Nov.
Abstract
The retroviral structural protein, Gag, is capable of independently assembling into virus-like particles (VLPs) in living cells and in vitro. Immature VLPs of human immunodeficiency virus type 1 (HIV-1) and of Rous sarcoma virus (RSV) are morphologically distinct when viewed by transmission electron microscopy (TEM). To better understand the nature of the Gag-Gag interactions leading to these distinctions, we constructed vectors encoding several RSV/HIV-1 chimeric Gag proteins for expression in either insect cells or vertebrate cells. We used TEM, confocal fluorescence microscopy, and a novel correlative scanning EM (SEM)-confocal microscopy technique to study the assembly properties of these proteins. Most chimeric proteins assembled into regular VLPs, with the capsid (CA) domain being the primary determinant of overall particle diameter and morphology. The presence of domains between matrix and CA also influenced particle morphology by increasing the spacing between the inner electron-dense ring and the VLP membrane. Fluorescently tagged versions of wild-type RSV, HIV-1, or murine leukemia virus Gag did not colocalize in cells. However, wild-type Gag proteins colocalized extensively with chimeric Gag proteins bearing the same CA domain, implying that Gag interactions are mediated by CA. A dramatic example of this phenomenon was provided by a nuclear export-deficient chimera of RSV Gag carrying the HIV-1 CA domain, which by itself localized to the nucleus but relocalized to the cytoplasm in the presence of wild type HIV-1 Gag. Wild-type and chimeric Gag proteins were capable of coassembly into a single VLP as viewed by correlative fluorescence SEM if, and only if, the CA domain was derived from the same virus. These results imply that the primary selectivity of Gag-Gag interactions is determined by the CA domain.
Figures
FIG. 1.
Schematic diagram and descriptions of wild-type and chimeric RSV/HIV-1 Gag proteins and summary of phenotype. RSV domains are in white; HIV-1 domains are in gray.
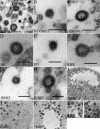
FIG. 2.
Thin-section TEM of wild-type and chimeric RSV/HIV-1 VLPs. (A) Pellet from mixed medium collected from SF9 cells expressing RSV and HIV-1 Gag. (B and C) Pellets from medium collected from DF1 cells expressing Gag proteins by transient transfection. (D to M) SF9 cells expressing Gag proteins by baculovirus infection. Wild-type RSV Gag (B); wild-type HIV-1 Gag (C); HR (D); RH (E); RHR (F); HRH2 (G); pellet from medium collected from SF9 cells expressing RHR2 (H); HRH (I); RnHc (J); HnRc (K); high magnification of RnHc (L); high magnification of HnRc (M). Scale bars, 100 nm.
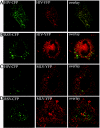
FIG. 3.
Fluorescence visualization of CFP or YFP labeled RSV, HIV-1, and MLV Gag in DF1 cells. HIV-1 Gag-CFP and HIV-1 Gag-YFP (A); RSV Gag-CFP and HIV-1 Gag-YFP (B); HIV-1 Gag-CFP and MLV Gag-YFP (C); RSV Gag-CFP and MLV Gag-YFP (D). Images are pseudocolored (CFP = green; YFP = red) and represent a single focal plane.
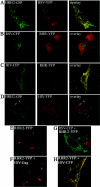
FIG. 4.
Fluorescence visualization of chimeric and wild type Gag proteins. Samples were prepared as in the results shown in Fig. 3. HRH2-CFP and RSV Gag-YFP (A); RSV Gag-CFP and RHR-YFP (B); HIV-1 Gag-CFP and RHR-YFP (C); HRH2-CFP and HIV-1 Gag-YFP (D); RHR2 alone (40× objective) (E); RHR2-YFP and nonlabeled HIV-1 Gag (40× objective) (F); RSV Gag-CFP and RHR2-YFP (G); RHR2-YFP and HIV-1 Gag-CFP (H). Images are pseudocolored (CFP = green, YFP = red) and represent a single focal plane.
FIG. 5.
Correlative SEM and confocal visualization of VLPs from fluorescent wild-type and chimeric Gag proteins from DF1 cells. (A) RSV Gag-YFP and RSV Gag-CFP. Black arrows point to VLPs in SEM images (B) HIV-1 Gag-CFP and RHR-YFP. (C) HIV-1 Gag-YFP and HRH2-CFP particles. The last panel in each row represents an overlay of the SEM and confocal images. Fluorescent images are pseudocolored (CFP = green, YFP = red) and represent a single focal plane.
Similar articles
- Characterization of Rous sarcoma virus Gag particles assembled in vitro.
Yu F, Joshi SM, Ma YM, Kingston RL, Simon MN, Vogt VM. Yu F, et al. J Virol. 2001 Mar;75(6):2753-64. doi: 10.1128/JVI.75.6.2753-2764.2001. J Virol. 2001. PMID: 11222698 Free PMC article. - Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus.
Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. Briggs JA, et al. J Mol Biol. 2006 Jan 6;355(1):157-68. doi: 10.1016/j.jmb.2005.10.025. Epub 2005 Nov 2. J Mol Biol. 2006. PMID: 16289202 - The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the p10 Domain in Assembly.
Schur FK, Dick RA, Hagen WJ, Vogt VM, Briggs JA. Schur FK, et al. J Virol. 2015 Oct;89(20):10294-302. doi: 10.1128/JVI.01502-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223638 Free PMC article. - The Assembly of HTLV-1-How Does It Differ from HIV-1?
Herrmann D, Meng S, Yang H, Mansky LM, Saad JS. Herrmann D, et al. Viruses. 2024 Sep 27;16(10):1528. doi: 10.3390/v16101528. Viruses. 2024. PMID: 39459862 Free PMC article. Review. - Ultrastructure and morphogenesis of human immunodeficiency virus.
Nakai M, Goto T. Nakai M, et al. J Electron Microsc (Tokyo). 1996 Aug;45(4):247-57. doi: 10.1093/oxfordjournals.jmicro.a023441. J Electron Microsc (Tokyo). 1996. PMID: 8888583 Review.
Cited by
- Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM. Obr M, et al. Nat Struct Mol Biol. 2024 Sep 6. doi: 10.1038/s41594-024-01390-8. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39242978 - The NTD-CTD intersubunit interface plays a critical role in assembly and stabilization of the HIV-1 capsid.
Yufenyuy EL, Aiken C. Yufenyuy EL, et al. Retrovirology. 2013 Mar 6;10:29. doi: 10.1186/1742-4690-10-29. Retrovirology. 2013. PMID: 23497318 Free PMC article. - The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag.
Murcia PR, Arnaud F, Palmarini M. Murcia PR, et al. J Virol. 2007 Feb;81(4):1762-72. doi: 10.1128/JVI.01859-06. Epub 2006 Nov 29. J Virol. 2007. PMID: 17135320 Free PMC article. - A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice.
Phillips JM, Murray PS, Murray D, Vogt VM. Phillips JM, et al. EMBO J. 2008 May 7;27(9):1411-20. doi: 10.1038/emboj.2008.71. Epub 2008 Apr 10. EMBO J. 2008. PMID: 18401344 Free PMC article. - Quantifying protein-protein interactions of peripheral membrane proteins by fluorescence brightness analysis.
Smith EM, Macdonald PJ, Chen Y, Mueller JD. Smith EM, et al. Biophys J. 2014 Jul 1;107(1):66-75. doi: 10.1016/j.bpj.2014.04.055. Biophys J. 2014. PMID: 24988342 Free PMC article.
References
- Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources