A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement - PubMed (original) (raw)
A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement
Masanori Yamasaki et al. Plant Cell. 2005 Nov.
Abstract
Maize (Zea mays subsp mays) was domesticated from teosinte (Z. mays subsp parviglumis) through a single domestication event in southern Mexico between 6000 and 9000 years ago. This domestication event resulted in the original maize landrace varieties, which were spread throughout the Americas by Native Americans and adapted to a wide range of environmental conditions. Starting with landraces, 20th century plant breeders selected inbred lines of maize for use in hybrid maize production. Both domestication and crop improvement involved selection of specific alleles at genes controlling key morphological and agronomic traits, resulting in reduced genetic diversity relative to unselected genes. Here, we sequenced 1095 maize genes from a sample of 14 inbred lines and chose 35 genes with zero sequence diversity as potential targets of selection. These 35 genes were then sequenced in a sample of diverse maize landraces and teosintes and tested for selection. Using two statistical tests, we identified eight candidate genes. Extended gene sequencing of these eight candidate loci confirmed that six were selected throughout the gene, and the remaining two exhibited evidence of selection in the 3' portion of each gene. The selected genes have functions consistent with agronomic selection for nutritional quality, maturity, and productivity. Our large-scale screen for artificial selection allows identification of genes of potential agronomic importance even when gene function and the phenotype of interest are unknown.
Figures
Figure 1.
Effect of Domestication and Plant Breeding on Genetic Diversity of Maize Genes. The colored circles represent different alleles. The shaded areas indicate bottleneck effects placed on all genes by the processes of domestication and improvement (plant breeding). Our model assumes that there will be three types of genes: neutral genes that show reduction of diversity by the general bottleneck effects, domestication genes in which diversity is greatly reduced by selection between the teosintes and landraces, and improvement genes in which diversity is greatly reduced by selection between the landraces and inbreds.
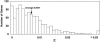
Figure 2.
Distribution of Genetic Diversity π for 1095 Maize Genes. The genes were sequenced in the panel of 14 diverse maize inbreds (Table 1).
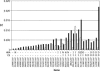
Figure 3.
The Genetic Diversity π in Landraces and Teosintes for Each of 35 Genes with Zero Diversity in Inbred Lines. The black bars indicate genetic diversity in the teosintes, and the white bars indicate genetic diversity in the landraces. Where no white bars are present, the genetic diversity in the landraces was zero. Inbreds are not shown as all are π = zero. The two rows under the figure indicate the test results for selection by HKA and CS analyses. The “-” indicates no significance; n = not tested in the HKA test due to inability to amplify a T. dactyloides orthologous sequence; I, improvement candidate; D, domestication candidate. The GenBank accession of the original Maize Mapping Project/DuPont unigene sequence is indicated under the bar.
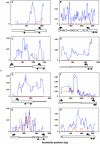
Figure 4.
Sliding-Window Analysis of the Genetic Diversity π in Maize Inbreds and Teosintes. For sliding-window analysis, π was calculated for segments of 100 bp at 10-bp intervals. Horizontal and vertical axes on the graphs indicate DNA sequence position and genetic diversity π, respectively. Red lines indicate genetic diversity in the inbreds, whereas blue lines indicate genetic diversity in the teosintes. For gene structure under the sliding-window graphs, white bars indicate the predicted exons, and black lines indicate introns or genomic regions. Left arrows and right arrows indicate the positions for predicted start codons and stop codons, respectively. The lines with two arrows under the gene structure indicate the sequencing regions in our initial screening. (A) AY108876; (B) AY107195; (C) AY110109; (D) AY105060; (E) AY108178; (F) AY106616; (G) AY107952; (H) AY106371.
Similar articles
- Genomic screening for artificial selection during domestication and improvement in maize.
Yamasaki M, Wright SI, McMullen MD. Yamasaki M, et al. Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review. - Patterns of selection and tissue-specific expression among maize domestication and crop improvement loci.
Hufford KM, Canaran P, Ware DH, McMullen MD, Gaut BS. Hufford KM, et al. Plant Physiol. 2007 Jul;144(3):1642-53. doi: 10.1104/pp.107.098988. Epub 2007 May 11. Plant Physiol. 2007. PMID: 17496114 Free PMC article. - Evidence of selection at the ramosa1 locus during maize domestication.
Sigmon B, Vollbrecht E. Sigmon B, et al. Mol Ecol. 2010 Apr;19(7):1296-311. doi: 10.1111/j.1365-294X.2010.04562.x. Epub 2010 Feb 24. Mol Ecol. 2010. PMID: 20196812 - The effects of artificial selection on the maize genome.
Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. Wright SI, et al. Science. 2005 May 27;308(5726):1310-4. doi: 10.1126/science.1107891. Science. 2005. PMID: 15919994 - Maize domestication and gene interaction.
Stitzer MC, Ross-Ibarra J. Stitzer MC, et al. New Phytol. 2018 Oct;220(2):395-408. doi: 10.1111/nph.15350. Epub 2018 Jul 23. New Phytol. 2018. PMID: 30035321 Review.
Cited by
- Genotype and haplotype analysis of the AZGP1 gene in cattle.
Zhang B, Guo YK, Li S, Zhang LZ, Lan XY, Hu SR, Chen H. Zhang B, et al. Mol Biol Rep. 2012 Dec;39(12):10475-9. doi: 10.1007/s11033-012-1928-x. Epub 2012 Oct 17. Mol Biol Rep. 2012. PMID: 23073769 - Evolution of neutral and flowering genes along pearl millet (Pennisetum glaucum) domestication.
Lakis G, Navascués M, Rekima S, Simon M, Remigereau MS, Leveugle M, Takvorian N, Lamy F, Depaulis F, Robert T. Lakis G, et al. PLoS One. 2012;7(5):e36642. doi: 10.1371/journal.pone.0036642. Epub 2012 May 14. PLoS One. 2012. PMID: 22606277 Free PMC article. - Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms.
Lu Y, Yan J, Guimarães CT, Taba S, Hao Z, Gao S, Chen S, Li J, Zhang S, Vivek BS, Magorokosho C, Mugo S, Makumbi D, Parentoni SN, Shah T, Rong T, Crouch JH, Xu Y. Lu Y, et al. Theor Appl Genet. 2009 Dec;120(1):93-115. doi: 10.1007/s00122-009-1162-7. Epub 2009 Oct 11. Theor Appl Genet. 2009. PMID: 19823800 - Comparative SNP diversity among four Eucalyptus species for genes from secondary metabolite biosynthetic pathways.
Külheim C, Yeoh SH, Maintz J, Foley WJ, Moran GF. Külheim C, et al. BMC Genomics. 2009 Sep 24;10:452. doi: 10.1186/1471-2164-10-452. BMC Genomics. 2009. PMID: 19775472 Free PMC article. - A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus).
Chapman MA, Pashley CH, Wenzler J, Hvala J, Tang S, Knapp SJ, Burke JM. Chapman MA, et al. Plant Cell. 2008 Nov;20(11):2931-45. doi: 10.1105/tpc.108.059808. Epub 2008 Nov 18. Plant Cell. 2008. PMID: 19017747 Free PMC article.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
- Bender, J. (2004). DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55, 41–68. - PubMed
- Bhattramakki, D., Dolan, M., Hanafey, M., Wineland, R., Vaske, D., Register III, J.C., Tingey, S.V., and Rafalski, A. (2002). Insertion-deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol. Biol. 48, 539–547. - PubMed
- Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources