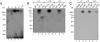Functional characterization of core promoter elements: the downstream core element is recognized by TAF1 - PubMed (original) (raw)
Functional characterization of core promoter elements: the downstream core element is recognized by TAF1
Dong-Hoon Lee et al. Mol Cell Biol. 2005 Nov.
Erratum in
- Mol Cell Biol. 2005 Dec;25(24):11192
Abstract
Downstream elements are a newly appreciated class of core promoter elements of RNA polymerase II-transcribed genes. The downstream core element (DCE) was discovered in the human beta-globin promoter, and its sequence composition is distinct from that of the downstream promoter element (DPE). We show here that the DCE is a bona fide core promoter element present in a large number of promoters and with high incidence in promoters containing a TATA motif. Database analysis indicates that the DCE is found in diverse promoters, supporting its functional relevance in a variety of promoter contexts. The DCE consists of three subelements, and DCE function is recapitulated in a TFIID-dependent manner. Subelement 3 can function independently of the other two and shows a TFIID requirement as well. UV photo-cross-linking results demonstrate that TAF1/TAF(II)250 interacts with the DCE subelement DNA in a sequence-dependent manner. These data show that downstream elements consist of at least two types, those of the DPE class and those of the DCE class; they function via different DNA sequences and interact with different transcription activation factors. Finally, these data argue that TFIID is, in fact, a core promoter recognition complex.
Figures
FIG. 1.
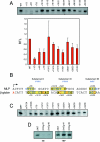
The adenovirus major late promoter contains a DCE type of downstream element. A. Scanning mutagenesis of the adenovirus MLP from +1 to +36. Triplet mutations from +1 to +36 were inserted into the Ad MLP templates and assayed by in vitro transcriptions using crude HeLa nuclear extracts. Transcription products were detected by primer extension. Relative transcription levels (RTL) represent the mutant transcription levels relative to the wild-type transcription level (WT). Mean values and standard deviations are calculated from the relative transcription levels (mutant versus wild type) from three to four experiments. Quantifications of the +22/24 and +34/36 mutants were approximately 80 to 90% of the wild-type MLP (n = 2) (data not shown). B. Sequence alignment of the wild-type human β-globin and adenovirus major late promoters. Boxed sequences, arrows, and numbering indicate positions of deleterious mutations in each promoter (panels A and C and reference 43). Bases in red indicate the positions of known β-thalassemia point mutations in the human β-globin promoter (2, 8, 24, 31, 57, 90), except for +13 (13). Blue-colored bases indicate the sequences representing DCE subelements. C. The β-globin DCE extends to positions +7/9 and does not show any erythroid cell specificity. Triplet mutations of the human β-globin promoter (43) were assayed by in vitro transcription using crude HeLa nuclear extracts. βWT, wild-type β-globin; 2.3GG, deleterious mutation in the human β-globin Inr element (44); mTATA, defective TATA box created by the replacement of the wild-type β-globin TATA box sequence with four TATA box β-thalassemia point mutations. D. Mutation of β-globin subelement II (βmSII) CTGT to AAAA. Two different preparations of a β-globin template containing mutations in all four base pairs of subelement II were assayed in an in vitro transcription assay using HeLa nuclear extracts (NE). The same templates were assayed using TBP in the reconstituted in vitro transcription assay as a negative control (see the text and Fig. 2).
FIG. 2.
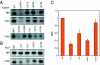
TFIID functionally recapitulates β-globin and MLP DCE activity in vitro. A. TFIID is necessary for β-globin DCE function. Using the highly purified in vitro transcription system, the three major β-globin DCE subelement mutants (Fig. 1B and C and reference 43) show decreased transcription using affinity-purified TFIID (two bottom panels) but not recombinant TBP (top panel). The mutant TATA box (mTATA) serves as a positive control. The second TFIID panel illustrates decreases in transcription using an Inr mutant (2,3GG), two mutations in SI (+7/9 and +10/12), and the SIII mutation (+31/33). WT and βWT, wild-type β-globin. B. MLP Inr mutant (+1/3) and DCE subelement mutants (Fig. 1A and B) show decreased transcription in vitro using the highly purified transcription system and affinity-purified TFIID but not recombinant TBP. C. Quantifications of the MLP TFIID-dependent transcriptions. Mean relative transcription levels (RTL) and standard deviations are calculated for three to four experiments.
FIG. 3.
DCE subelement III can function independently of subelements I and II. A. HSV UL38 wild-type (WT) and mutant promoter sequences. Predicted SIII sequences are in blue, and mutated bases are in red (26). In vitro transcriptions of the UL38 WT promoter and DAS mutants 1 and 3 (das1 and das3) were performed using HeLa nuclear extracts (NE) or a highly purified transcription system containing either TBP or affinity-purified TFIID. Below the in vitro transcription are quantifications of two sets of the in vitro transcription experiments. B. Adenovirus promoter sequences containing DCE sequences. Shown are several promoters from adenovirus 5 (GenBank accession numbers BK000408 and X02996). The promoters are indicated on the left. The arrow indicates the +1 transcriptional start site. The vertical boxes indicate positions that fit the Inr consensus sequence (YYANA/TYY [73]). The three boxes and blue lettering indicate, in the case of the MLP, experimentally derived positions of the three DCE subelements (Fig. 1). In the case of the remaining promoters, the boxes and blue lettering indicate predicted DCE subelements. C. In vitro transcription analysis of the wild-type adenovirus E3 promoter and a mutation of the predicted SIII (E3 mAGC; AGC is mutated to TTT). Shown are transcriptions using HeLa nuclear extracts (NE) or using the highly purified reconstituted transcription system (RTS) containing either recombinant TBP or affinity-purified TFIID. D. Titration of the adenovirus IX promoter containing either the wild-type promoter sequences (IX) or a mutation of the predicted SIII AGC sequence (IX mAGC; AGC is mutated to TTT). Shown are transcriptions using HeLa nuclear extracts. E. DCE subelement III function has severe spatial constraints. Wild-type and mutant E3 in vitro transcriptions using HeLa nuclear extracts are compared to the deletion or insertion of 3 base pairs between the subelement III AGC and the start site of transcription (−3AGC and +3AGC, respectively; the insertion of AAA was at +24 for +3AGC, and deletion of the TCG sequence was at +21 to +23 for −3AGC). F. Analysis of Ad MLP spacing mutants. The Ad MLP SIII was moved 4 bp closer to SII to establish a 10-bp interval between SII and SIII as in the β-globin promoter (−4SIII; the GGCC from +28 to +31 was deleted). In this context, SIII was also mutated to TTT (−4mSIII). Shown are results of duplicate in vitro transcriptions (using HeLa NE) of these two templates, compared to the wild-type Ad MLP (MLP).
FIG. 4.
TAF1/TAFII250 photo-cross-linking is observed with wild-type but not mutant MLP DCE subelements in the adenovirus MLP. A. Photo-cross-linking adducts were placed at the indicated positions relative to the transcriptional start site on the template strand of the wild-type adenovirus MLP and incubated with affinity-purified TFIID and rTFIIA (see Materials and Methods). PAGE and autoradiography were performed on the resulting cross-linked products. B. Photo-cross-linking adducts were placed at positions +9, +19, and +31 of the template strand of the adenovirus MLP wild-type or mutant DCE subelements (WT SI, mSI, etc.) The mutant subelement sequences are identical to those used in Fig. 1A. PAGE and autoradiography were performed on the resulting cross-linked products, with (lanes 2, 3, 5, 6, 8, 9) or without (lanes 1, 4, and 7) irradiation with UV light. Binding reactions were as described for panel A. C. TFIIA/TFIID photo-cross-linked products (at positions +9, +19, and +31 of the template strand) were dissociated and immunoprecipitated with either protein A-agarose beads alone (lane 1), an anti-FLAG antibody (αFLAG; Sigma) plus protein A-agarose beads (lanes 2, 4, and 6), or an anti-TAF1/TAFII250 antibody plus protein A-agarose beads (lanes 3, 5, and 7). The immunoprecipitates were then analyzed by PAGE and autoradiography.
FIG. 5.
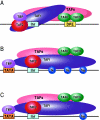
Models depicting the interaction of TFIID with the DPE and DCE. In the three models, TAF1 and TAF2 are jointly responsible for Inr element recognition (for a review, see reference 73). A. DPE sequence recognition is established by TAF6/TAF9 components of TFIID. This interaction results in a unique TFIID conformation that in turn results in the formation of a DPE-specific PIC (PICDPE). B. TFIID interacts with the DCE subelements via TAF1. Again, this results in a TFIID conformation that is different than a TFIID/DPE interaction. This also leads to the formation of a DCE-specific PIC (PICDCE). C. A similar interaction of TFIID occurs with promoters containing only SIII of the DCE. In all three cases, these unique PICs consist of their own unique set of factors and cofactors that, in the end, manifest themselves as different regulatory phenomena.
Similar articles
- SAGA mediates transcription from the TATA-like element independently of Taf1p/TFIID but dependent on core promoter structures in Saccharomyces cerevisiae.
Watanabe K, Kokubo T. Watanabe K, et al. PLoS One. 2017 Nov 27;12(11):e0188435. doi: 10.1371/journal.pone.0188435. eCollection 2017. PLoS One. 2017. PMID: 29176831 Free PMC article. - TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter.
Wright KJ, Marr MT 2nd, Tjian R. Wright KJ, et al. Proc Natl Acad Sci U S A. 2006 Aug 15;103(33):12347-52. doi: 10.1073/pnas.0605499103. Epub 2006 Aug 8. Proc Natl Acad Sci U S A. 2006. PMID: 16895980 Free PMC article. - Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription.
Xu M, Gonzalez-Hurtado E, Martinez E. Xu M, et al. Biochim Biophys Acta. 2016 Apr;1859(4):553-63. doi: 10.1016/j.bbagrm.2016.01.005. Epub 2016 Jan 26. Biochim Biophys Acta. 2016. PMID: 26824723 Free PMC article. - The DPE, a core promoter element for transcription by RNA polymerase II.
Kadonaga JT. Kadonaga JT. Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review. - TRF2: TRansForming the view of general transcription factors.
Zehavi Y, Kedmi A, Ideses D, Juven-Gershon T. Zehavi Y, et al. Transcription. 2015;6(1):1-6. doi: 10.1080/21541264.2015.1004980. Epub 2015 Jan 14. Transcription. 2015. PMID: 25588059 Free PMC article. Review.
Cited by
- Conserved DNA sequence features underlie pervasive RNA polymerase pausing.
Gajos M, Jasnovidova O, van Bömmel A, Freier S, Vingron M, Mayer A. Gajos M, et al. Nucleic Acids Res. 2021 May 7;49(8):4402-4420. doi: 10.1093/nar/gkab208. Nucleic Acids Res. 2021. PMID: 33788942 Free PMC article. - Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae.
Sugihara F, Kasahara K, Kokubo T. Sugihara F, et al. Nucleic Acids Res. 2011 Jan;39(1):59-75. doi: 10.1093/nar/gkq741. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805245 Free PMC article. - Rrn7 protein, an RNA polymerase I transcription factor, is required for RNA polymerase II-dependent transcription directed by core promoters with a HomolD box sequence.
Rojas DA, Moreira-Ramos S, Zock-Emmenthal S, Urbina F, Contreras-Levicoy J, Käufer NF, Maldonado E. Rojas DA, et al. J Biol Chem. 2011 Jul 29;286(30):26480-6. doi: 10.1074/jbc.M111.224337. Epub 2011 Jun 14. J Biol Chem. 2011. PMID: 21673110 Free PMC article. - Three novel downstream promoter elements regulate MHC class I promoter activity in mammalian cells.
Lee N, Iyer SS, Mu J, Weissman JD, Ohali A, Howcroft TK, Lewis BA, Singer DS. Lee N, et al. PLoS One. 2010 Dec 13;5(12):e15278. doi: 10.1371/journal.pone.0015278. PLoS One. 2010. PMID: 21179443 Free PMC article. - The punctilious RNA polymerase II core promoter.
Vo Ngoc L, Wang YL, Kassavetis GA, Kadonaga JT. Vo Ngoc L, et al. Genes Dev. 2017 Jul 1;31(13):1289-1301. doi: 10.1101/gad.303149.117. Genes Dev. 2017. PMID: 28808065 Free PMC article. Review.
References
- Abmayr, S., J. Workman, and R. Roeder. 1988. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 2:542-553. - PubMed
- Athanassiadou, A., A. Papachatzopoulou, N. Zoumbos, G. M. Maniatis, and R. Gibbs. 1994. A novel beta-thalassaemia mutation in the 5′ untranslated region of the beta-globin gene. Br. J. Haematol. 88:307-310. - PubMed
- Bucher, P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563-578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM064844/GM/NIGMS NIH HHS/United States
- Z01 DK060001/ImNIH/Intramural NIH HHS/United States
- K01 DK 60001/DK/NIDDK NIH HHS/United States
- GM 64844/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources