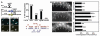LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans - PubMed (original) (raw)
LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans
Andrew S Yoo et al. Science. 2005.
Abstract
Cell-cell interactions and cross-talk between signaling pathways specify Caenorhabditis elegans vulval precursor cells (VPCs) to adopt a spatial pattern: a central "1 degrees " VPC, in which epidermal growth factor receptor (EGFR)-mitogen-activated protein kinase (MAPK) activity is high and LIN-12/Notch activity is low, flanked by two "2 degrees " VPCs, in which LIN-12/Notch activity is high and EGFR-MAPK activity is low. Here, we identify a microRNA gene, mir-61, as a direct transcriptional target of LIN-12 and show that expression of mir-61 promotes the 2 degrees fate. We also identify vav-1, the ortholog of the Vav oncogene, as a target of mir-61, and show that down-regulation of VAV-1 promotes lin-12 activity in specifying the 2 degrees fate. Our results suggest that lin-12, mir-61, and vav-1 form a feedback loop that helps maximize lin-12 activity in the presumptive 2 degrees VPCs.
Figures
Fig. 1
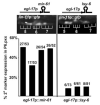
mir-61 is a direct target of LIN-12 in presumptive 2° VPCs. (A) An inductive signal from the anchor cell (AC) of the gonad activates EGFR-MAPK signaling primarily in P6.p, and a lateral signal from P6.p activates LIN-12 in P5.p and P7.p. The descendants of the 1° and 2° VPCs form the vulva, and the progeny of 3° VPCs fuse with the hypodermal syncytium. We used the 1° fate marker _arIs92[egl-17_p::cfp-lacZ] (3), the 2° fate marker nIs106[lin-11_p::gfp] (21), and the 3° fate marker arIs101[K09H11.1_p::yfp] (22). (B) mir-61 is expressed in P5.p and P7.p. The mir-61 promoter contains two LBSs that are conserved in the C. briggsae ortholog of mir-61 (5). A reporter containing 1 kb upstream of mir-61 fused to YFP is expressed in P5.p and P7.p (22) and their daughters (shown here). Prominent expression in cells of the gonad in which LIN-12 is active is also seen. (C) LBSs are required for mir-61 expression in P5.p and P7.p. Two LBSs (YRTGRGAA) (3, 23) that are conserved in C. briggsae were mutated to YRAGRGAA; a third nonconserved sequence, RTGGGAA, was also mutated to RAGGGAA. In three individual lines analyzed, expression of YFP in P5.p and P7.p disappeared. Expression in cells in which lin-12 is not known to play a role in cell fate specification was normal, but gonadal expression was also abolished.
Fig. 2
Ectopic expression of mir-61 in P6.p confers 2° fate characteristics. Expression of the 2° fate marker _nIs106[lin-11_p::gfp] (see Fig. 1A) was assessed when egl-17_p (3, 24) was used for ectopic expression of mir-61 or the unrelated miRNA lsy-6 (25) in P6.p and its descendants. Ectopic lin11_p::gfp expression in P6.p descendants was observed only when mir-61 was expressed. Each bar represents an independent transgenic line. The number of individuals expressing the 2° marker out of the total is given above each bar.
Fig. 3
vav-1 is a target of mir-61. (A) Rapid assay to validate predicted miRNA targets. Components are expressed in coelomocytes to determine whether a miRNA causes down-regulation of YFP in a sensor construct containing a test UTR without affecting CFP in a marker construct with the neutral unc-54 3′ UTR (26). Here, an array carrying unc-122_p::mir-61_ was combined with an array carrying the unc-122_p::cfp::unc-54 3 ′_UTR marker and hlh-8_p::yfp::vav-1 3 ′_UTR sensor (right) or an array carrying hlh-8_p::yfp::unc-54 3_ ′UTR in lieu of the sensor (left) (6). The presence of the _mir-61_–expressing array does not affect expression of YFP produced from a sensor construct that contains a nontarget UTR (in this case, the unc-54 UTR) (triangles). In contrast, YFP expressed from a sensor construct that contains a target UTR (shown here, the vav-1 UTR) is not seen, whereas the CFP marker shows that the array is present and expressed (arrows). The graphs indicate the percentage of worms that show the down-regulation of YFP signal. The alignment shows predicted configuration of mir-61 (in red) binding to its target site in the 3′ UTR of vav-1 (in blue). (B) VAV-1 is posttranscriptionally down-regulated in P5.p and P7.p. The 8.4-kb upstream region of vav-1 drives expression of YFP in all VPCs and their daughters. When the unc-54 3′ UTR is replaced by vav-1 3′ UTR, down-regulation of YFP expression in P5.px and P7.px is evident. Mutation of the _mir-61_–complementary sequence in the vav-1 3′UTR from TAGTCA to GTCGAC causes persistent YFP expression. In the graph, each bar represents an individual line. **P < 0.01 by Fisher’s exact test. We minimized the potential lack of expression in P5.px or P7.px due to genetic mosaicism by including data only for animals in which expression could be seen in P3.px, P4.px, and P8.px.
Fig. 4
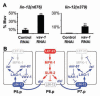
vav-1 is a negative regulator of lin-12 activity in the VPCs. (A) vav-1(RNAi) enhances lin-12 activity in VPCs. The average percentage of worms with a Multivulva (Muv) phenotype (three or more pseudovulvae) on three independent plates is shown. For lin-12(n676); vav-1(RNAi), the number of Muv hermaphrodites out of the total per plate was 50 out of 109, 49 out of 112, and 54 out of 125, with the control lin-12(n676); gfp(RNAi) values of 8 out of 157, 17 out of 168, and 18 out of 152. For lin-12(n379); vav-1(RNAi), the numbers were 20 out of 187, 25 out of 208, and 13 out of 167, with lin-12(n379); gfp(RNAi) values of 1 out of 204, 1 out of 228, and 2 out of 225. Error bar indicates SD. **P < 0.01 by Student’s t test. (B) mir-61, vav-1, and the circuitry underlying specification of the 2° VPC fate. Activation of the EGFR-MAPK pathway in P6.p has two consequences for lateral signaling: transcription of the three Delta/Serrate/LAG-2 (DSL) ligands that constitute the lateral signal, which activates LIN-12/Notch in P5.p and P7.p (27), and internalization of LIN-12, which is necessary for lateral signal activity (28). Activation of LIN-12 in P5.p and P7.p activates a set of lst genes that counteract the EGFR-MAPK pathway (3), and mir-61, which posttranscriptionally down-regulates VAV-1 to promote lin-12 activity.
Comment in
- Developmental biology. Encountering microRNAs in cell fate signaling.
Karp X, Ambros V. Karp X, et al. Science. 2005 Nov 25;310(5752):1288-9. doi: 10.1126/science.1121566. Science. 2005. PMID: 16311325 No abstract available.
Similar articles
- Developmental biology. Encountering microRNAs in cell fate signaling.
Karp X, Ambros V. Karp X, et al. Science. 2005 Nov 25;310(5752):1288-9. doi: 10.1126/science.1121566. Science. 2005. PMID: 16311325 No abstract available. - Integration of EGFR and LIN-12/Notch Signaling by LIN-1/Elk1, the Cdk8 Kinase Module, and SUR-2/Med23 in Vulval Precursor Cell Fate Patterning in Caenorhabditis elegans.
Underwood RS, Deng Y, Greenwald I. Underwood RS, et al. Genetics. 2017 Dec;207(4):1473-1488. doi: 10.1534/genetics.117.300192. Epub 2017 Sep 27. Genetics. 2017. PMID: 28954762 Free PMC article. - Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development.
Yoo AS, Bais C, Greenwald I. Yoo AS, et al. Science. 2004 Jan 30;303(5658):663-6. doi: 10.1126/science.1091639. Science. 2004. PMID: 14752159 - Vulval development.
Sternberg PW. Sternberg PW. WormBook. 2005 Jun 25:1-28. doi: 10.1895/wormbook.1.6.1. WormBook. 2005. PMID: 18050418 Free PMC article. Review. - LIN-12/Notch signaling in C. elegans.
Greenwald I. Greenwald I. WormBook. 2005 Aug 8:1-16. doi: 10.1895/wormbook.1.10.1. WormBook. 2005. PMID: 18050403 Free PMC article. Review.
Cited by
- ALG-1, a microRNA argonaute, promotes vulva induction in C. elegans.
Zhen S, Rocheleau CE. Zhen S, et al. MicroPubl Biol. 2024 Oct 15;2024:10.17912/micropub.biology.001373. doi: 10.17912/micropub.biology.001373. eCollection 2024. MicroPubl Biol. 2024. PMID: 39493436 Free PMC article. - A lineage-resolved cartography of microRNA promoter activity in C. elegans empowers multidimensional developmental analysis.
Xu W, Liu J, Qi H, Si R, Zhao Z, Tao Z, Bai Y, Hu S, Sun X, Cong Y, Zhang H, Fan D, Xiao L, Wang Y, Li Y, Du Z. Xu W, et al. Nat Commun. 2024 Mar 30;15(1):2783. doi: 10.1038/s41467-024-47055-4. Nat Commun. 2024. PMID: 38555276 Free PMC article. - Identifying the Caenorhabditis elegans vulval transcriptome.
Zhang Q, Hrach H, Mangone M, Reiner DJ. Zhang Q, et al. G3 (Bethesda). 2022 May 30;12(6):jkac091. doi: 10.1093/g3journal/jkac091. G3 (Bethesda). 2022. PMID: 35551383 Free PMC article. - SALSA, a genetically encoded biosensor for spatiotemporal quantification of Notch signal transduction in vivo.
Shaffer JM, Greenwald I. Shaffer JM, et al. Dev Cell. 2022 Apr 11;57(7):930-944.e6. doi: 10.1016/j.devcel.2022.03.008. Dev Cell. 2022. PMID: 35413239 Free PMC article. - Plasma miR-21 as a potential predictor in prediabetic individuals with a positive family history of type 2 diabetes mellitus.
Yazdanpanah Z, Kazemipour N, Kalantar SM, Vahidi Mehrjardi MY. Yazdanpanah Z, et al. Physiol Rep. 2022 Jan;10(2):e15163. doi: 10.14814/phy2.15163. Physiol Rep. 2022. PMID: 35076188 Free PMC article.
References
- Sternberg PW. WormBook. 2005. www.wormbook.org. - PMC - PubMed
- Greenwald I. WormBook. 2005. www.wormbook.org. - PubMed
- Yoo AS, Bais C, Greenwald I. Science. 2004;303:663. - PubMed
- Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843. - PubMed
- Supporting Online Material is available on Science Online.
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA095389-03/CA/NCI NIH HHS/United States
- CA095389/CA/NCI NIH HHS/United States
- R01 CA095389-02/CA/NCI NIH HHS/United States
- R01 CA095389/CA/NCI NIH HHS/United States
- R01 CA095389-05/CA/NCI NIH HHS/United States
- R01 CA095389-06A1/CA/NCI NIH HHS/United States
- R01 CA095389-01A1/CA/NCI NIH HHS/United States
- R01 CA095389-04/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous