Expansion of the BioCyc collection of pathway/genome databases to 160 genomes - PubMed (original) (raw)
Expansion of the BioCyc collection of pathway/genome databases to 160 genomes
Peter D Karp et al. Nucleic Acids Res. 2005.
Abstract
The BioCyc database collection is a set of 160 pathway/genome databases (PGDBs) for most eukaryotic and prokaryotic species whose genomes have been completely sequenced to date. Each PGDB in the BioCyc collection describes the genome and predicted metabolic network of a single organism, inferred from the MetaCyc database, which is a reference source on metabolic pathways from multiple organisms. In addition, each bacterial PGDB includes predicted operons for the corresponding species. The BioCyc collection provides a unique resource for computational systems biology, namely global and comparative analyses of genomes and metabolic networks, and a supplement to the BioCyc resource of curated PGDBs. The Omics viewer available through the BioCyc website allows scientists to visualize combinations of gene expression, proteomics and metabolomics data on the metabolic maps of these organisms. This paper discusses the computational methodology by which the BioCyc collection has been expanded, and presents an aggregate analysis of the collection that includes the range of number of pathways present in these organisms, and the most frequently observed pathways. We seek scientists to adopt and curate individual PGDBs within the BioCyc collection. Only by harnessing the expertise of many scientists we can hope to produce biological databases, which accurately reflect the depth and breadth of knowledge that the biomedical research community is producing.
Figures
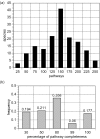
Figure 1
Distribution of BioCyc pathways across species. (a) Frequency analysis: the _x_-axis shows the number of detected pathways and the _y_-axis the number of species containing those pathways. (b) Completeness analysis: the _x_-axis shows the percentage of pathway completeness and the _y_-axis the frequency of pathways with the corresponding degree of completeness—more than 60% of pathways are more than 50% complete in the BioCyc collection of PGDBs.
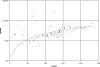
Figure 2
Relationship between number of pathways (_x_-axis) and number of genes (_y_-axis) for all species in the BioCyc collection. Bacterial species are shown in light grey, archaeal species in open-grey squares and eukaryotes in black. The fitted line—a linear regression curve—refers to Bacteria only; most Archaea exhibit a similar relationship. The two outlier bacterial species with fewer than 25 pathways can be seen on the left part of the graph: Mycobacterium avium paratuberculosis and Ralstonia solanacearum GMI1000. The three largest eukaryotic genomes with >10 000 genes show a significant underrepresentation of pathways for their genome size.
Similar articles
- The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases.
Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD. Caspi R, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D473-9. doi: 10.1093/nar/gkp875. Epub 2009 Oct 22. Nucleic Acids Res. 2010. PMID: 19850718 Free PMC article. - The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases.
Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. Caspi R, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D623-31. doi: 10.1093/nar/gkm900. Epub 2007 Oct 27. Nucleic Acids Res. 2008. PMID: 17965431 Free PMC article. - The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases.
Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Pujar A, Shearer AG, Travers M, Weerasinghe D, Zhang P, Karp PD. Caspi R, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D742-53. doi: 10.1093/nar/gkr1014. Epub 2011 Nov 18. Nucleic Acids Res. 2012. PMID: 22102576 Free PMC article. - DOOR: a prokaryotic operon database for genome analyses and functional inference.
Cao H, Ma Q, Chen X, Xu Y. Cao H, et al. Brief Bioinform. 2019 Jul 19;20(4):1568-1577. doi: 10.1093/bib/bbx088. Brief Bioinform. 2019. PMID: 28968679 Review. - The challenge of constructing, classifying, and representing metabolic pathways.
Caspi R, Dreher K, Karp PD. Caspi R, et al. FEMS Microbiol Lett. 2013 Aug;345(2):85-93. doi: 10.1111/1574-6968.12194. Epub 2013 Jun 27. FEMS Microbiol Lett. 2013. PMID: 23746312 Free PMC article. Review.
Cited by
- Learning virulent proteins from integrated query networks.
Cadag E, Tarczy-Hornoch P, Myler PJ. Cadag E, et al. BMC Bioinformatics. 2012 Dec 2;13:321. doi: 10.1186/1471-2105-13-321. BMC Bioinformatics. 2012. PMID: 23198735 Free PMC article. - Correlation-Based Network Analysis of Metabolite and Enzyme Profiles Reveals a Role of Citrate Biosynthesis in Modulating N and C Metabolism in Zea mays.
Toubiana D, Xue W, Zhang N, Kremling K, Gur A, Pilosof S, Gibon Y, Stitt M, Buckler ES, Fernie AR, Fait A. Toubiana D, et al. Front Plant Sci. 2016 Jul 12;7:1022. doi: 10.3389/fpls.2016.01022. eCollection 2016. Front Plant Sci. 2016. PMID: 27462343 Free PMC article. - NaviCell Web Service for network-based data visualization.
Bonnet E, Viara E, Kuperstein I, Calzone L, Cohen DP, Barillot E, Zinovyev A. Bonnet E, et al. Nucleic Acids Res. 2015 Jul 1;43(W1):W560-5. doi: 10.1093/nar/gkv450. Epub 2015 May 9. Nucleic Acids Res. 2015. PMID: 25958393 Free PMC article. - Algorithms for modeling global and context-specific functional relationship networks.
Zhu F, Panwar B, Guan Y. Zhu F, et al. Brief Bioinform. 2016 Jul;17(4):686-95. doi: 10.1093/bib/bbv065. Epub 2015 Aug 6. Brief Bioinform. 2016. PMID: 26254431 Free PMC article. - The Edinburgh human metabolic network reconstruction and its functional analysis.
Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I. Ma H, et al. Mol Syst Biol. 2007;3:135. doi: 10.1038/msb4100177. Epub 2007 Sep 18. Mol Syst Biol. 2007. PMID: 17882155 Free PMC article.
References
- Romero P., Karp P. PseudoCyc, a pathway-genome database for Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 2003;5:230–239. - PubMed
- Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. - PMC - PubMed
- Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. - PMC - PubMed