Autophagy: molecular machinery for self-eating - PubMed (original) (raw)
Review
Autophagy: molecular machinery for self-eating
T Yorimitsu et al. Cell Death Differ. 2005 Nov.
Abstract
Autophagy is a highly conserved process in eukaryotes in which the cytoplasm, including excess or aberrant organelles, is sequestered into double-membrane vesicles and delivered to the degradative organelle, the lysosome/vacuole, for breakdown and eventual recycling of the resulting macromolecules. This process has an important role in various biological events such as adaptation to changing environmental conditions, cellular remodeling during development and differentiation, and determination of lifespan. Auto-phagy is also involved in preventing certain types of disease, although it may contribute to some pathologies. Recent studies have identified many components that are required to drive this complicated cellular process. Auto-phagy-related genes were first identified in yeast, but homologs are found in all eukaryotes. Analyses in a range of model systems have provided huge advances toward understanding the molecular basis of autophagy. Here we review our current knowledge on the machinery and molecular mechanism of autophagy.
Figures
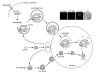
Figure 1
Autophagy and the Cvt pathway in yeast. Morphological steps are schematically shown in autophagy and the Cvt pathway. Both pathways require the sequestration of cargo components within distinct double-membrane vesicles, whose formation is thought to occur at the PAS. Autophagy is induced by inactivation of Tor kinase under starvation conditions such as nitrogen depletion. Organelles and bulk cytoplasm are sequestered into a double-membrane autophagosome, which is 300-900 nm in diameter. The Cvt pathway is a biosynthetic process that delivers resident hydrolases including Ape1 to the vacuole in vegetative conditions. The Cvt vesicle (140-160 nm in diameter) enwraps specific cargos such as the prApe1 oligomer (Ape1 complex). The Ape1 complex is also sequestered specifically by autophagosomes under starvation conditions. Once completed, the double-membrane vesicles target to and fuse with the vacuole, and the single-membrane vesicles (autophagic or Cvt body) are released into the lumen. Subsequently, these vesicles are degraded, allowing maturation of prApe1 into the mature form (mApe1) by removal of its propeptide, and the degradation of the cytoplasm. Fluorescent images show that the cargo protein CFP-Ape1 localizes at the perivacuolar PAS (white arrowhead) marked by YFP-Atg8; the vacuole is labeled with FM 4-64 in the merged image. DIC, differential interference contrast
Figure 2
Regulatory complex for autophagy induction. Tor kinase regulates the induction of autophagy upon sensing the nutrient conditions. Atg1 kinase, which is essential for both autophagy and the Cvt pathway, forms a complex with several proteins that are characterized as being preferentially involved in the Cvt pathway (in white circles) or autophagy (in dark gray circle). Under nutrient-rich conditions, Tor kinase is active, and Atg13 is hyperphosphorylated. This highly phosphorylated Atg13 has a lower affinity for Atg1 and Atg17, and autophagy is downregulated. Under starvation conditions, Tor is inactive and Atg13 is hypophosphorylated. Dephosphorylated Atg13 interacts with Atg1 and Atg17 with a higher affinity. The enhancement of the formation of the Atg1-Atg13-Atg17 complex mediates the induction of autophagy. The Atg20-Atg24 complex, Atg11, and Vac8 might also belong to this complex, but a complete holo-complex as depicted has not been identified
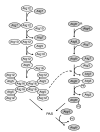
Figure 3
Selective recognition and packaging of cargo, and vesicle formation in the Cvt pathway. The Cvt pathway is an Atg process that allows the selective transport of the vacuolar hydrolases Ape1 and Ams1 into the vacuole. The preApe1 is synthesized in the cytosol, subsequently assembles into dodecamers and further forms a larger oligomer, termed the Ape1 complex. The receptor protein Atg19 binds to the Ape1 complex through the prApe1 propeptide to form the Cvt complex; the Ams1 oligomer is recruited to the complex via its binding to Atg19. Atg11 is assembled with the Cvt complex by interacting with Atg19 and guides the complex to the PAS. Atg11 forms a homodimer or homo-oligomer at the PAS, although it is not clear whether it occurs after or before the arrival at this site. Once the Cvt complex reaches the PAS, Atg components such as Atg8 conjugated to phosphatidylethanolamine (Atg8-PE) are recruited for vesicle formation. Atg19 further interacts with Atg8-PE, which might ensure the proper packaging of the cargo into the forming Cvt vesicle. Atg11 dissociates from the Cvt complex at the PAS prior to vesicle completion, although the exact timing is unknown
Figure 4
Two conjugation machineries. One conjugation begins with Atg7, which is homologous to the E1 ubiquitin-activating enzyme, linking to Atg12 through the formation of a thioester bond involving the C-terminal glycine residue of Atg12. Activated Atg12 is transferred to the E2-like protein Atg10, which catalyzes the conjugation with Atg5 through the formation of an isopeptide bond between the activated glycine of Atg12 and an internal lysine of Atg5. Finally, Atg16 is assembled with the Atg12-Atg5 conjugate. Atg16 can also form a homotetramer, resulting in a multimer of the Atg12-Atg5-Atg16 complex. The other system starts with the cysteine protease Atg4, which proteolytically removes the C-terminal arginine of Atg8 to allow access to a glycine residue. The same E1-like Atg7 activates this exposed glycine residue and then transfers activated Atg8 to Atg3. Finally, Atg3 mediates the conjugation of Atg8 with phosphatidylethanolamine to form Atg8-PE. The Atg12-Atg5-Atg16 conjugate is also required for stability of Atg8-PE, although the mechanism remains unclear. Atg12-Atg5-Atg16 and Atg8-PE both localize to the PAS and appear to be involved in vesicle formation. Atg8-PE is a structural component of the completed Cvt vesicle and autophagosome, but is enriched on the inner membrane. During or soon after vesicle formation, Atg8-PE on the vesicle outer membrane is cleaved off by Atg4 protease
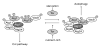
Figure 5
Two PtdIns 3-kinase complexes in yeast. (a) Both complexes share Vps15, Vps30/Atg6 and Vps34. Complex I has Atg14 and functions in autophagy and the Cvt pathway. Complex II contains Vps38 and is involved in the CPY and Mvb pathways. (b) It is hypothesized that complex I might generate PtdIns(3)P for autophagy and the Cvt pathway at the PAS. The Atg20-Atg24 complex, Atg18, Atg21 and Atg27 are recruited to the PAS through their association with PtdIns(3)P
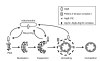
Figure 6
Recycling of Atg9 and Atg23. Atg9 and Atg23 localize at the PAS and at multiple peripheral punctate structures (shown to be the mitochondria for Atg9). Retrieval of the integral membrane protein Atg9 from the PAS involves the Atg1-Atg13 complex, Atg2, and Atg18. The PtdIns 3-kinase complex I is proposed to generate PtdIns(3)P at the PAS, and Atg18 is likely localized to the PAS through its association with this phosphoinositide. The Atg1-Atg13 complex induces the interaction of Atg2, Atg9 and Atg18. Finally, this ternary complex allows Atg9 retrieval from the PAS. Atg23 is a peripheral membrane protein. It is not known whether Atg23 is recruited to the PAS from a cytosolic or membrane-associated pool. In contrast to Atg9, a high level of Atg1 kinase activity is needed to mediate the retrieval of Atg23 from the PAS
Figure 7
Schematic model for the step of vesicle formation. First, membrane nucleation is dependent on the PtdIns 3-kinase complex I, and Atg9. Atg proteins, including the Atg12-Atg5-Atg16 complex and Atg8-PE conjugate, components of the vesicle-forming machinery, are recruited to the PAS. The Atg12-Atg5-Atg16 complex mediates the expansion of the vesicle possibly by acting as a coat. Atg8-PE is recruited into the forming vesicle dependent on the Atg12-Atg5-Atg16 complex, and also acts in the elongation of the vesicle as a structural component. Atg9 cycles between the PAS and mitochondria; Atg9 cycling may supply the lipid for the expanding membrane. Upon, or just prior to, vesicle completion, the coat proteins and the Atg proteins, except for the Atg8-PE that is oriented toward the lumen (and Atg19), are dissociated from the vesicle. Finally, the completed vesicle can fuse with the vacuole
Similar articles
- Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells.
Kim J, Klionsky DJ. Kim J, et al. Annu Rev Biochem. 2000;69:303-42. doi: 10.1146/annurev.biochem.69.1.303. Annu Rev Biochem. 2000. PMID: 10966461 Review. - An overview of the molecular mechanism of autophagy.
Yang Z, Klionsky DJ. Yang Z, et al. Curr Top Microbiol Immunol. 2009;335:1-32. doi: 10.1007/978-3-642-00302-8_1. Curr Top Microbiol Immunol. 2009. PMID: 19802558 Free PMC article. Review. - The molecular mechanism of autophagy.
Wang CW, Klionsky DJ. Wang CW, et al. Mol Med. 2003 Mar-Apr;9(3-4):65-76. Mol Med. 2003. PMID: 12865942 Free PMC article. Review. - Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy.
Wurmser AE, Emr SD. Wurmser AE, et al. J Cell Biol. 2002 Aug 19;158(4):761-72. doi: 10.1083/jcb.200112050. Epub 2002 Aug 19. J Cell Biol. 2002. PMID: 12186856 Free PMC article. - The late stages of autophagy: how does the end begin?
Noda T, Fujita N, Yoshimori T. Noda T, et al. Cell Death Differ. 2009 Jul;16(7):984-90. doi: 10.1038/cdd.2009.54. Epub 2009 May 8. Cell Death Differ. 2009. PMID: 19424283 Review.
Cited by
- The dual role of autophagy in suppressing and promoting hepatocellular carcinoma.
Mohammed WH, Sulaiman GM, Abomughaid MM, Klionsky DJ, Abu-Alghayth MH. Mohammed WH, et al. Front Cell Dev Biol. 2024 Oct 11;12:1472574. doi: 10.3389/fcell.2024.1472574. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39463763 Free PMC article. Review. - The influence of circular RNAs on autophagy and disease progression.
Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan Q, Tang Y, He Y, Liao Q, Li X, Wu X, Xiang B, Zhou M, Li Y, Li G, Li X, Zeng Z, Guo C, Xiong W. Wang Y, et al. Autophagy. 2022 Feb;18(2):240-253. doi: 10.1080/15548627.2021.1917131. Epub 2021 Apr 27. Autophagy. 2022. PMID: 33904341 Free PMC article. Review. - BNIP3 is degraded by ULK1-dependent autophagy via MTORC1 and AMPK.
Park CW, Hong SM, Kim ES, Kwon JH, Kim KT, Nam HG, Choi KY. Park CW, et al. Autophagy. 2013 Mar;9(3):345-60. doi: 10.4161/auto.23072. Epub 2013 Jan 4. Autophagy. 2013. PMID: 23291726 Free PMC article. - Atorvastatin suppresses the progression of cervical cancer via regulation of autophagy.
Sheng B, Song Y, Zhang J, Li R, Wang Z, Zhu X. Sheng B, et al. Am J Transl Res. 2020 Sep 15;12(9):5252-5268. eCollection 2020. Am J Transl Res. 2020. PMID: 33042417 Free PMC article. - Endoplasmic reticulum stress in cardiometabolic disorders.
Ozcan L. Ozcan L. Curr Atheroscler Rep. 2012 Oct;14(5):469-75. doi: 10.1007/s11883-012-0270-z. Curr Atheroscler Rep. 2012. PMID: 22847769 Review.
References
- Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2435–2444. - PubMed
- Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2420–2434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases