Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function - PubMed (original) (raw)
Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function
Courtney J Haycraft et al. PLoS Genet. 2005 Oct.
Abstract
Intraflagellar transport (IFT) proteins are essential for cilia assembly and have recently been associated with a number of developmental processes, such as left-right axis specification and limb and neural tube patterning. Genetic studies indicate that IFT proteins are required for Sonic hedgehog (Shh) signaling downstream of the Smoothened and Patched membrane proteins but upstream of the Glioma (Gli) transcription factors. However, the role that IFT proteins play in transduction of Shh signaling and the importance of cilia in this process remain unknown. Here we provide insights into the mechanism by which defects in an IFT protein, Tg737/Polaris, affect Shh signaling in the murine limb bud. Our data show that loss of Tg737 results in altered Gli3 processing that abrogates Gli3-mediated repression of Gli1 transcriptional activity. In contrast to the conclusions drawn from genetic analysis, the activity of Gli1 and truncated forms of Gli3 (Gli3R) are unaffected in Tg737 mutants at the molecular level, indicating that Tg737/Polaris is differentially involved in specific activities of the Gli proteins. Most important, a negative regulator of Shh signaling, Suppressor of fused, and the three full-length Gli transcription factors localize to the distal tip of cilia in addition to the nucleus. Thus, our data support a model where cilia have a direct role in Gli processing and Shh signal transduction.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
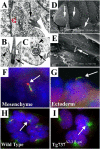
Figure 1. Cilia Are Present on Both Mesenchymal and Ectodermal Cells of the Developing Limb
(A–C) Transmission electron micrographs of limb bud mesenchyme show cilia (arrows) closely associated with the Golgi apparatus (“G”). The cilia exhibited a 9 + 0 structure (C) and are often found in deep depressions in the membranes (B). Frequently, small vesicles are observed fusing or budding with the surrounding membrane (arrowheads in [B] and [C]). (D and E) Scanning electron micrographs of the limb ectoderm show a single cilium (arrows) on nearly all ectodermal cells. (F and G) Immunolocalization of Polaris (red) and acetylated α-tubulin (green) in frozen sections of limb buds shows that Polaris concentrates at the base and tip of the axoneme in both mesenchymal (F) and ectodermal (G) cells. Nuclei are blue. (H and I) In primary cultures of cells isolated from E11.5 limb buds, cilia (arrow in H) are also present when visualized with anti-acetylated α-tubulin (green) and anti-Polaris (red) antisera (H). Cilia are absent on cells isolated from Tg737 Δ2–3β-gal mutant limb buds (I); however, the stabilized microtubules were still evident around the basal body region (arrow). The nuclear staining for Polaris is present in the Tg737 Δ2–3β-gal cells, indicating that it is nonspecific. Nuclei are blue.
Figure 2. Shh Signaling Is Defective in Tg737 Δ2–3β-gal Mutants
(A and B) In situ hybridization analysis of Ptch1 (A) and Gli1 (B) expression indicates that they are not expressed in the posterior limb buds of Tg737 Δ2–3β-gal mutant embryos (E10.5; right panels) as they are in wild-type controls (E10.5, left panels). (C) Incubation of wild-type limb bud cells with ShhN-CM results in upregulation of Gli1 and Ptch1 expression (left lanes) compared to vector conditioned medium, whereas no increase is seen in cells isolated from Tg737 Δ2–3β-gal mutant limb buds (right lanes). The relative levels of induction standardized to actin are indicated below each lane.
Figure 3. Gli2 and Full-Length Gli3 Function Is Disrupted in Tg737 Δ2–3β-gal Mutant Cells
(A) Infection of primary limb bud cells (E11.5) with Gli1::GFP expressing adenovirus induces increased Ptch1 transcription in wild-type cells when compared to infection with GFP-only virus (GFP). Coinfection of wild-type cells with Gli1::GFP and Gli3::GFP results in a decrease in the level of Ptch1 expression when compared to cells infected with Gli1::GFP only. As seen in wild-type cells, infection of Tg737 Δ2–3β-gal mutants with Gli1::GFP induced Ptch1 expression. However, full-length Gli3::GFP was unable to suppress Gli1::GFP-mediated induction of Ptch1 in the absence of Polaris (Tg737 Δ2–3β-gal). No expression was seen in controls without reverse transcriptase (−RT). (B) Infection of wild-type cells with a Gli2::GFP expressing adenovirus induced Ptch1 expression; however, in Tg737 Δ2–3β-gal primary limb bud cells, infection with the Gli2::GFP expressing adenovirus failed to induce the pathway, when compared to infection with GFP-only virus (GFP, right lanes). (C) Western blot analysis of proteins isolated from whole E11.5 wild-type embryos (left lane) shows that Gli3 is predominantly found in the processed repressor form (Gli3R). While some Gli3R is evident in the mutant samples, a large proportion of Gli3 remains unprocessed (Gli3A) in Tg737 Δ2–3β-gal mutants (right lane). (D) Coinfection of wild-type or Tg737 Δ2–3β-gal mutant cells with Gli1::GFP and a truncated Gli3R::GFP indicates that Gli3R is able to repress Gli1-mediated induction of Ptch1. Numbers below each lane in (A), (B), and (D) indicate the expression level of Ptch1 relative to the actin control for the experiment shown.
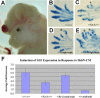
Figure 4. Gli3Xt-J/+;Tg737orpk/orpk Embryos Resemble Gli3Xt-J/Xt-J Null Embryos and Are Responsive to ShhN-CM
(A) Example of exencephaly observed in Gli3Xt-J/+;Tg737orpk/orpk embryos that is never observed in Gli3Xt-J/ + or Tg737orpk/orpk mutants alone. (B–E) Functional interaction of Gli3 and Tg737 in digit development. Whereas Gli3Xt-J/+ (C) and Tg737orpk/orpk (D) embryos each develop one extra preaxial digit (asterisks), Gli3Xt-J/+;Tg737orpk/orpk embryos (E) develop multiple ectopic digits compared to wild-type embryos (B). Anterior is to the top. (F) Incubation of cells from Gli3+/+;Tg737+/+, Gli3Xt-J/+;Tg737+/+, and Gli3Xt-J/+;Tg737orpk/orpk mutant mice with ShhN-CM resulted in increased expression of Gli1 when compared to cells from the same embryo treated with control medium, as determined by quantitative RT-PCR analysis. The results are reported for four littermates of the indicated genotypes. Each sample was analyzed in duplicate, and results are reported as the average fold increase. Unlike Tg737 Δ2–3β-gal (null) mutants, which are nonresponsive to ShhN-CM, the Gli3Xt-J/+;Tg737orpk/orpk samples are able to respond and activate the pathway, indicating that the Gli3Xt-J/+;Tg737orpk/orpk phenotype does not resemble that of Tg737 Δ2–3β-gal (null) mutants but rather that of Gli3Xt-J/ Xt-J.
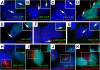
Figure 5. GFP-Tagged Gli Proteins Localize to the Distal Tip of the Cilium in Primary Limb Cell Cultures
(A–D) Cells were isolated from limb buds of wild-type embryos at E11.5 and infected with the indicated adenovirus. All three full-length GFP-tagged Gli proteins (green) localize to a domain in the cilium axoneme, which is visualized with anti-acetylated α-tubulin staining (red). In contrast, Gli3R::GFP is restricted to the nucleus and is not detected in this domain (D). (E) The full-length Gli::GFP proteins (Gli2::GFP shown here) do not colocalize with the basal body at the base of the cilium, which is visualized with anti-γ-tubulin staining (red), indicating that the full-length Gli proteins localize to the tips of the cilia. (F and G) Gli2::GFP (F) and Gli3::GFP (G) colocalize with a subdomain of Polaris (red) at the distal tip of the cilium. (H–K) In Tg737 Δ2–3β-gal mutant limb bud cells, the GFP-tagged Gli1 (H), Gli2 (I), and Gli3 (J) proteins localize to the nucleus and in the region of stabilized microtubules around the MTOC marked by anti-acetylated α-tubulin. In contrast, the processed form of Gli3 (Gli3R::GFP) (K) is detected only in the nucleus. Insets in all panels show the GFP (green) and nuclear (blue) staining only for the indicated cilium (arrow) or region (box).
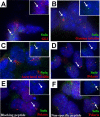
Figure 6. Endogenous Sufu and Gli3 Localize to the Distal Tip of the Cilium in Wild-Type Primary Limb Cell Cultures
Sufu (green) and endogenous Gli3 (red) concentrate in the same domain in cultured wild-type limb bud cells (A). As shown for the full-length Gli::GFP proteins, endogenous Sufu does not colocalize with γ-tubulin (red) (B), but is concentrated in a domain at the distal end of the acetylated α-tubulin staining (red) (C). Sufu also partially overlaps with a domain of Polaris (red) (D) in cultured wild-type limb bud cells. Pre-incubation of anti-Sufu antiserum with the immunizing peptide (E), but not with a nonspecific peptide (F), blocks staining at the distal tip of the cilium (anti-Polaris, red; anti-Sufu, green). Inset in all panels shows Gli3 (A) or Sufu (B–F) staining only in the indicated cilium (arrow).
Similar articles
- Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli.
May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. May SR, et al. Dev Biol. 2005 Nov 15;287(2):378-89. doi: 10.1016/j.ydbio.2005.08.050. Epub 2005 Oct 17. Dev Biol. 2005. PMID: 16229832 - Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors.
Liu A, Wang B, Niswander LA. Liu A, et al. Development. 2005 Jul;132(13):3103-11. doi: 10.1242/dev.01894. Epub 2005 Jun 1. Development. 2005. PMID: 15930098 - Unique and complimentary activities of the Gli transcription factors in Hedgehog signaling.
Lipinski RJ, Gipp JJ, Zhang J, Doles JD, Bushman W. Lipinski RJ, et al. Exp Cell Res. 2006 Jul 1;312(11):1925-38. doi: 10.1016/j.yexcr.2006.02.019. Epub 2006 Mar 29. Exp Cell Res. 2006. PMID: 16571352 - Essential roles of Gli3 and sonic hedgehog in pattern formation and developmental anomalies caused by their dysfunction.
Motoyama J. Motoyama J. Congenit Anom (Kyoto). 2006 Sep;46(3):123-8. doi: 10.1111/j.1741-4520.2006.00114.x. Congenit Anom (Kyoto). 2006. PMID: 16922918 Review. - Gli Proteins: Regulation in Development and Cancer.
Niewiadomski P, Niedziółka SM, Markiewicz Ł, Uśpieński T, Baran B, Chojnowska K. Niewiadomski P, et al. Cells. 2019 Feb 11;8(2):147. doi: 10.3390/cells8020147. Cells. 2019. PMID: 30754706 Free PMC article. Review.
Cited by
- Ulk4 promotes Shh signaling by regulating Stk36 ciliary localization and Gli2 phosphorylation.
Zhou M, Han Y, Jiang J. Zhou M, et al. Elife. 2023 Dec 14;12:RP88637. doi: 10.7554/eLife.88637. Elife. 2023. PMID: 38096226 Free PMC article. - Primary cilia and kidney injury: current research status and future perspectives.
Wang S, Dong Z. Wang S, et al. Am J Physiol Renal Physiol. 2013 Oct 15;305(8):F1085-98. doi: 10.1152/ajprenal.00399.2013. Epub 2013 Jul 31. Am J Physiol Renal Physiol. 2013. PMID: 23904226 Free PMC article. Review. - Review of literature: genes related to postaxial polydactyly.
Verma PK, El-Harouni AA. Verma PK, et al. Front Pediatr. 2015 Feb 11;3:8. doi: 10.3389/fped.2015.00008. eCollection 2015. Front Pediatr. 2015. PMID: 25717468 Free PMC article. Review. - Systematic analysis of cilia characteristics and Hedgehog signaling in five immortal cell lines.
Gómez AE, Christman AK, Van De Weghe JC, Finn M, Doherty D. Gómez AE, et al. PLoS One. 2022 Dec 29;17(12):e0266433. doi: 10.1371/journal.pone.0266433. eCollection 2022. PLoS One. 2022. PMID: 36580465 Free PMC article. - Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans.
Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Mukhopadhyay S, et al. EMBO J. 2007 Jun 20;26(12):2966-80. doi: 10.1038/sj.emboj.7601717. Epub 2007 May 17. EMBO J. 2007. PMID: 17510633 Free PMC article.
References
- Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. - PubMed
- Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, et al. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. - PubMed
- Moyer JH, Lee-Tischler MJ, Kwon HY, Schrick JJ, Avner ED, et al. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. - PubMed
- Yoder BK, Tousson A, Millican L, Wu JH, Bugg CE, Jr, et al. Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am J Physiol Renal Physiol. 2002;282:F541–F552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases