Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study - PubMed (original) (raw)
Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study
A Terao et al. Neuroscience. 2006.
Abstract
Previous studies have demonstrated that macromolecular synthesis in the brain is modulated in association with the occurrence of sleep and wakefulness. Similarly, the spectral composition of electroencephalographic activity that occurs during sleep is dependent on the duration of prior wakefulness. Since this homeostatic relationship between wake and sleep is highly conserved across mammalian species, genes that are truly involved in the electroencephalographic response to sleep deprivation might be expected to be conserved across mammalian species. Therefore, in the rat cerebral cortex, we have studied the effects of sleep deprivation on the expression of immediate early gene and heat shock protein mRNAs previously shown to be upregulated in the mouse brain in sleep deprivation and in recovery sleep after sleep deprivation. We find that the molecular response to sleep deprivation and recovery sleep in the brain is highly conserved between these two mammalian species, at least in terms of expression of immediate early gene and heat shock protein family members. Using Affymetrix Neurobiology U34 GeneChips , we also screened the rat cerebral cortex, basal forebrain, and hypothalamus for other genes whose expression may be modulated by sleep deprivation or recovery sleep. We find that the response of the basal forebrain to sleep deprivation is more similar to that of the cerebral cortex than to the hypothalamus. Together, these results suggest that sleep-dependent changes in gene expression in the cerebral cortex are similar across rodent species and therefore may underlie sleep history-dependent changes in sleep electroencephalographic activity.
Figures
Figure 1
Hourly total sleep time values (mean ± S.E.M.) in male Wistar rats (n=13) over a 32 h period: 24 h baseline followed by 6 h of SD and a subsequent 2 h recovery period. Black horizontal bars indicate dark periods, and white bars indicate light period. Asterisk denotes significant difference (*p < 0.05) between experimental and corresponding control group by Student-Newman-Keuls post-hoc test followed by one-way ANOVA.
Figure 2
Expression of egr3, fra2, grp78 and grp94 mRNA in the rat cerebral cortex across the four experimental conditions determined by real-time RT-PCR. g3pdh expression was used as an internal standard. Values are mean ± S.E.M. P values are based on ANOVA; asterisks denote significant differences (*p < 0.05) between experimental and corresponding control group by Tukey-Kramer post-hoc test. Abbreviations: C, control; R, recovery sleep; SD, sleep deprived; ZT, zeitgeber time; NS, no significant difference.
Figure 3
Expression of arc mRNA in mouse and rat cerebral cortex after 6 h SD and after recovery from SD. A. Autoradiographs of Northern blots of total RNA from mouse cerebral cortex (upper panel) and rat cerebral cortex (lower panel) hybridized to [32P]-labeled arc and β-actin probes. Based on OD260 readings, approximately 5 µg total RNA was loaded in each lane. Abbreviations: SD, animals deprived of sleep for 6 h; C, control animals sacrificed 6 h after lights on (ZT6); R, animals allowed 2 h (rat) or 4 h (mouse) recovery sleep after 6 h SD; C’, control mice sacrificed 10 h after lights on (ZT10; upper panel) or control rats sacrificed eight hours after lights-on (ZT8; lower panel). Graph based on optical density ratios from resultant autoradiographs; values are mean ± S.E.M. B. Taqman analysis of arc expression in mouse (upper panel) or rat (lower panel) cortex. P values are based on ANOVA; asterisks denote significant differences (*p < 0.05) between experimental and corresponding control group by Tukey-Kramer post-hoc test.
Figure 4
Comparison of normalized intensity values obtained in SD vs. ZT6 control (left column) and RS vs. ZT8 control (right column) comparisons for the cerebral cortex (A), basal forebrain (B), and hypothalamus (C). For each gene, the mean signal intensity value for the experimental group (SD or RS; y-axis) is plotted against the mean of the corresponding control group (control; x-axis). The dotted lines in each graph demarcate the threshold for 1.5-fold upregulation or downregulation in the experimental group relative to the control group. Accordingly, points that lie above the upper dotted line in each graph represent genes that underwent an increase in expression more than 1.5-fold relative to control values; points that lie below the lower dotted line underwent a decrease in expression more than 1.5-fold relative to control values. Values for which there was a qualitative difference between experimental and control groups (i.e., present in control chips and absent in experimental chips, or vice versa) are not plotted. Abbreviations: BF=basal forebrain; Cx=cerebral cortex; Hy=hypothalamus.
Figure 5
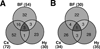
Venn diagrams illustrating the number of genes up-regulated (A) or down-regulated (B) at least 1.5-fold in each of the three brain regions at ZT6 during SD. Abbreviations: BF=basal forebrain; Cx=cerebral cortex; Hy=hypothalamus.
Figure 6
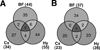
Venn diagrams illustrating the number of genes up-regulated (A) or down-regulated (B) at least 1.5-fold in each of the three brain regions at ZT8 during RS. Abbreviations: BF=basal forebrain; Cx=cerebral cortex; Hy=hypothalamus.
Figure 7
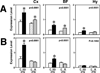
Expression of nr4a3 (A) and ngfi-b (B) mRNAs in the cerebral cortex (Cx), basal forebrain (BF), and hypothalamus (Hy) across the four experimental conditions. g3pdh expression was used as an internal standard. Values are mean ± S.E.M. P values are based on the ANOVA; asterisks denote significant differences (*p < 0.05) between experimental and corresponding control group by Tukey-Kramer post-hoc test. Abbreviations: C, control; R, recovery sleep; SD, sleep deprived; ZT, Zeitgeber time.
Similar articles
- Region-specific changes in immediate early gene expression in response to sleep deprivation and recovery sleep in the mouse brain.
Terao A, Greco MA, Davis RW, Heller HC, Kilduff TS. Terao A, et al. Neuroscience. 2003;120(4):1115-24. doi: 10.1016/s0306-4522(03)00395-6. Neuroscience. 2003. PMID: 12927216 - Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep.
Terao A, Steininger TL, Hyder K, Apte-Deshpande A, Ding J, Rishipathak D, Davis RW, Heller HC, Kilduff TS. Terao A, et al. Neuroscience. 2003;116(1):187-200. doi: 10.1016/s0306-4522(02)00695-4. Neuroscience. 2003. PMID: 12535952 - How sleep deprivation affects gene expression in the brain: a review of recent findings.
Cirelli C. Cirelli C. J Appl Physiol (1985). 2002 Jan;92(1):394-400. doi: 10.1152/jappl.2002.92.1.394. J Appl Physiol (1985). 2002. PMID: 11744682 Review. - Modulation of brain gene expression during sleep and wakefulness: a review of recent findings.
Tononi G, Cirelli C. Tononi G, et al. Neuropsychopharmacology. 2001 Nov;25(5 Suppl):S28-35. doi: 10.1016/S0893-133X(01)00322-0. Neuropsychopharmacology. 2001. PMID: 11682270 Review.
Cited by
- Proteomic profiling of the rat cerebral cortex in sleep and waking.
Cirelli C, Pfister-Genskow M, McCarthy D, Woodbury R, Tononi G. Cirelli C, et al. Arch Ital Biol. 2009 Sep;147(3):59-68. Arch Ital Biol. 2009. PMID: 20014652 Free PMC article. - Sex-specific regulation of the cortical transcriptome in response to sleep deprivation.
Shi T, Shah I, Dang Q, Taylor L, Jagannath A. Shi T, et al. Front Neurosci. 2024 Mar 5;17:1303727. doi: 10.3389/fnins.2023.1303727. eCollection 2023. Front Neurosci. 2024. PMID: 38504908 Free PMC article. - Sleep and oligodendrocyte functions.
Bellesi M. Bellesi M. Curr Sleep Med Rep. 2015 Mar 1;1(1):20-26. doi: 10.1007/s40675-014-0008-2. Curr Sleep Med Rep. 2015. PMID: 25821717 Free PMC article. - Primed to Sleep: The Dynamics of Synaptic Plasticity Across Brain States.
Seibt J, Frank MG. Seibt J, et al. Front Syst Neurosci. 2019 Feb 1;13:2. doi: 10.3389/fnsys.2019.00002. eCollection 2019. Front Syst Neurosci. 2019. PMID: 30774586 Free PMC article. - Effects of sleep and wake on oligodendrocytes and their precursors.
Bellesi M, Pfister-Genskow M, Maret S, Keles S, Tononi G, Cirelli C. Bellesi M, et al. J Neurosci. 2013 Sep 4;33(36):14288-300. doi: 10.1523/JNEUROSCI.5102-12.2013. J Neurosci. 2013. PMID: 24005282 Free PMC article.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
- Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. In: Kryger MH, et al., editors. Principles and Practice of Sleep Medicine. Philadelphia: W.B. Saunders; 2000. pp. 377–390.
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL/MH59658/HL/NHLBI NIH HHS/United States
- R01 HL059658-08/HL/NHLBI NIH HHS/United States
- R01 HL059658-06/HL/NHLBI NIH HHS/United States
- R01 HL059658/HL/NHLBI NIH HHS/United States
- R01 HL059658-07/HL/NHLBI NIH HHS/United States
- R01 HL059658-09/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources