Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase - PubMed (original) (raw)
. 2005 Dec;17(12):3301-10.
doi: 10.1105/tpc.105.034645. Epub 2005 Oct 28.
Affiliations
- PMID: 16258034
- PMCID: PMC1315370
- DOI: 10.1105/tpc.105.034645
Free PMC article
Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase
Sang Yeol Kim et al. Plant Cell. 2005 Dec.
Free PMC article
Abstract
Winter-annual accessions of Arabidopsis thaliana are often characterized by a requirement for exposure to the cold of winter to initiate flowering in the spring. The block to flowering prior to cold exposure is due to high levels of the flowering repressor FLOWERING LOCUS C (FLC). Exposure to cold promotes flowering through a process known as vernalization that epigenetically represses FLC expression. Rapid-cycling accessions typically have low levels of FLC expression and therefore do not require vernalization. A screen for mutants in which a winter-annual Arabidopsis is converted to a rapid-cycling type has identified a putative histone H3 methyl transferase that is required for FLC expression. Lesions in this methyl transferase, EARLY FLOWERING IN SHORT DAYS (EFS), result in reduced levels of histone H3 Lys 4 trimethylation in FLC chromatin. EFS is also required for expression of other genes in the FLC clade, such as MADS AFFECTING FLOWERING2 and FLOWERING LOCUS M. The requirement for EFS to permit expression of several FLC clade genes accounts for the ability of efs lesions to suppress delayed flowering due to the presence of FRIGIDA, autonomous pathway mutations, or growth in noninductive photoperiods. efs mutants exhibit pleiotropic phenotypes, indicating that the role of EFS is not limited to the regulation of flowering time.
Figures
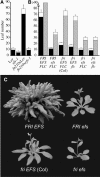
Figure 1.
efs Mutations Suppress the Late-Flowering Phenotype of FRI. (A) Allelism tests between efs-1 in the L_er_ background and fn210 in the _FRI_-Col background. The closed portion of the bars indicates the number of rosette leaves formed by the primary shoot apical meristem prior to flowering. The open portion of the bars indicates the number of cauline leaves. Error bars indicate 1
sd
. (B) Number of leaves formed prior to flowering for the indicated genotypes. Black and gray bars represent plants grown in LD and SD, respectively. Error bars indicate 1
sd
. (C) The effect of efs mutations on flowering time in _FRI_-Col background (top) or Col background (bottom). Plants were grown in LD.
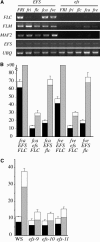
Figure 2.
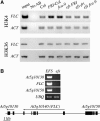
Effect of EFS on Gene Expression and Flowering Time of Autonomous Pathway Mutants. (A) RT-PCR analysis of flowering time gene expression in wild-type and efs mutant backgrounds. RNA was isolated from 14-d-old seedlings grown in LD. Tissue was harvested 4 h after lights on. UBIQUITIN (UBQ) was used as a control for loading. (B) and (C) Effect of efs on the flowering time of fca and fve mutants (B) and the effect of efs alleles in the Ws genetic background (C). The closed portion of the bar indicates the number of rosette leaves formed by the primary shoot apical meristem prior to flowering, while the open portion indicates the number of cauline leaves. Black and gray bars represent plants grown in LD and SD, respectively. Error bars indicate 1
sd
.
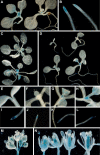
Figure 3.
Histochemical Analysis of Gene Expression. To minimize variation in GUS staining, plants that appear in the same panel were stained in parallel. Unless otherwise mentioned, all plants were grown in LD and are in the _FRI_-Col background. (A) and (B) FLC:GUS expression in shoots and roots in the wild-type _FRI_-Col (left) or efs mutant background (right). (C) SOC1:GUS expression in wild-type _FRI_-Col (top) or efs mutant background (bottom). (D) EFS:GUS expression during vegetative development. Plants were harvested 2 (top left), 4 (top right), 6 (bottom left), and 8 DAG. (E) to (L) Higher magnification images of the shoot apex and root apex of the plants shown in (D). 2 DAG ([E] and [I]), 4 DAG ([F] and [J]), 6 DAG ([G] and [K]), and 8 DAG ([H] and [L]). (M) EFS:GUS expression in the inflorescence. (N) EFS:GUS expression in flowers.
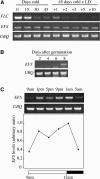
Figure 4.
The EFS Gene and Predicted Protein. (A) Schematic representation of the EFS gene (top; black boxes indicate exons) and protein (bottom). Positions of lesions in efs mutants are indicated. (B) Clustal alignment of the SET, pre-SET, and post-SET domains of EFS, ASH1, SET1, and SET2. Identical residues are shaded in dark gray and similar residues in light gray.
Figure 5.
RT-PCR Analysis of EFS Expression. (A) EFS and FLC expression during vernalization. Eight-day-old seedlings were exposed to cold for 0, 15, 30, or 45 d and harvested for RNA isolation. Also, seedlings exposed to cold for 45 d were grown at 22°C for 1, 2, 3, 5, or 10 d prior to harvest. (B) EFS expression during vegetative development. Shoots were harvested at 2, 4, 6, and 8 DAG. (C) Daily fluctuations in EFS expression. Eight-day-old seedlings were harvested at the indicated times. UBQ was used as a control for loading in all experiments.
Figure 6.
efs Mutations Affect Histone H3-K4 Trimethylation and Gene Expression around the FLC Locus. (A) Chromatin immunoprecipitation analysis of histone H3-K4 trimethylation and H3-K36 dimethylation state of FLC chromatin in efs and related lines. The input is Col chromatin before immunoprecipitation. “No AB” refers to the control sample lacking the antitrimethyl H3-K4 or antidimethyl H3-K36 antibody. ACTIN (ACT) served as an internal control. (B) RT-PCR analysis of genes flanking FLC in wild-type _FRI_-Col and efs mutant backgrounds. UBQ was used as a control for loading.
Figure 7.
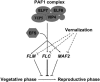
Model of Activation of the FLC Clade Genes by EFS. EFS protein is recruited to the FLC clade loci by the PAF-like complex and presumably methylates H3-K4, resulting in activating expression of these genes. Vernalization represses expression of the FLC clade genes to accelerate floral transition. Lines with arrows indicate upregulation/activation of gene expression, and lines with bars indicate repression.
Similar articles
- SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis.
Kim SY, Michaels SD. Kim SY, et al. Development. 2006 Dec;133(23):4699-707. doi: 10.1242/dev.02684. Epub 2006 Nov 1. Development. 2006. PMID: 17079264 - The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms.
Jean Finnegan E, Kovac KA, Jaligot E, Sheldon CC, James Peacock W, Dennis ES. Jean Finnegan E, et al. Plant J. 2005 Nov;44(3):420-32. doi: 10.1111/j.1365-313X.2005.02541.x. Plant J. 2005. PMID: 16236152 - Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3.
Sung S, Amasino RM. Sung S, et al. Nature. 2004 Jan 8;427(6970):159-64. doi: 10.1038/nature02195. Nature. 2004. PMID: 14712276 - Role of chromatin modification in flowering-time control.
He Y, Amasino RM. He Y, et al. Trends Plant Sci. 2005 Jan;10(1):30-5. doi: 10.1016/j.tplants.2004.11.003. Trends Plant Sci. 2005. PMID: 15642521 Review. - Vernalization: winter and the timing of flowering in plants.
Kim DH, Doyle MR, Sung S, Amasino RM. Kim DH, et al. Annu Rev Cell Dev Biol. 2009;25:277-99. doi: 10.1146/annurev.cellbio.042308.113411. Annu Rev Cell Dev Biol. 2009. PMID: 19575660 Review.
Cited by
- Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14.
Ning YQ, Ma ZY, Huang HW, Mo H, Zhao TT, Li L, Cai T, Chen S, Ma L, He XJ. Ning YQ, et al. Nucleic Acids Res. 2015 Feb 18;43(3):1469-84. doi: 10.1093/nar/gku1382. Epub 2015 Jan 10. Nucleic Acids Res. 2015. PMID: 25578968 Free PMC article. - Histone Lysine-to-Methionine Mutations Reduce Histone Methylation and Cause Developmental Pleiotropy.
Sanders D, Qian S, Fieweger R, Lu L, Dowell JA, Denu JM, Zhong X. Sanders D, et al. Plant Physiol. 2017 Apr;173(4):2243-2252. doi: 10.1104/pp.16.01499. Epub 2017 Feb 15. Plant Physiol. 2017. PMID: 28202597 Free PMC article. - Arabidopsis trithorax-related3/SET domain GROUP2 is required for the winter-annual habit of Arabidopsis thaliana.
Yun JY, Tamada Y, Kang YE, Amasino RM. Yun JY, et al. Plant Cell Physiol. 2012 May;53(5):834-46. doi: 10.1093/pcp/pcs021. Epub 2012 Feb 28. Plant Cell Physiol. 2012. PMID: 22378382 Free PMC article. - Embryonic epigenetic reprogramming by a pioneer transcription factor in plants.
Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y. Tao Z, et al. Nature. 2017 Nov 2;551(7678):124-128. doi: 10.1038/nature24300. Epub 2017 Oct 25. Nature. 2017. PMID: 29072296 - The Arabidopsis thaliana SET-domain-containing protein ASHH1/SDG26 interacts with itself and with distinct histone lysine methyltransferases.
Valencia-Morales Mdel P, Camas-Reyes JA, Cabrera-Ponce JL, Alvarez-Venegas R. Valencia-Morales Mdel P, et al. J Plant Res. 2012 Sep;125(5):679-92. doi: 10.1007/s10265-012-0485-7. Epub 2012 Mar 22. J Plant Res. 2012. PMID: 22438063
References
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. - PubMed
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. - PubMed
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29, 4319–4333. - PMC - PubMed
- Beisel, C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. (2002). Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419, 857–862. - PubMed
- Bezerra, I.C., Michaels, S.D., Schomburg, F.M., and Amasino, R.M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40, 112–119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases