Histone H3.3 deposition at E2F-regulated genes is linked to transcription - PubMed (original) (raw)
Histone H3.3 deposition at E2F-regulated genes is linked to transcription
Laetitia Daury et al. EMBO Rep. 2006 Jan.
Abstract
The histone variant H3.3 can be incorporated in chromatin independently of DNA synthesis. By imaging using green fluorescent protein-tagged histones, H3.3 deposition has been found to be linked with transcriptional activation. Here, we investigated H3.3 incorporation during G1 progression on cell-cycle-regulated E2F-dependent genes and on some control loci. We transiently transfected resting cells with an expression vector for tagged H3.3 and we analysed its presence by chromatin immunoprecipitation. We found that replication-independent H3.3 deposition occurred on actively transcribed genes, but not on silent loci, thereby confirming its link with transcription. Interestingly, we observed similar levels of H3.3 occupancy on promoters and on the coding regions of the corresponding genes, indicating that H3.3 deposition is not restricted to promoters. Finally, H3.3 occupancy correlated with the presence of transcription-competent RNA polymerase II. Taken together, our results support the hypothesis that H3.3 is incorporated after disruption of nucleosomes mediated by transcription elongation.
Figures
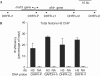
Figure 1
Histone H3 is not depleted from the dihydrofolate reductase promoter during G1 progression. (A) Schematic representation of the mouse dhfr (dihydrofolate reductase) locus, with the position of probes detected by quantitative PCR (Q-PCR) in chromatin immunoprecipitation (ChIP) experiments. Note the presence of the msh3 gene 600 bp upstream of the dhfr transcription start site and transcribed in the opposite direction. (B) NIH3T3 cells were starved of serum for 48 h and then induced for 8 h with 20% FCS. Cells were then subjected to a ChIP assay using an anti-H3 antibody or no antibody as a control (NA). The amounts of the indicated sequences were measured by Q-PCR. A representative experiment out of three is shown. Ab, antibody.
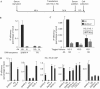
Figure 2
Study of histone H3.3 deposition during G1 progression. (A) Description of the experimental procedure. NIH3T3 cells (400,000 cells/10 cm dish, six dishes per immunoprecipitation) were starved in 0.5% FCS for 48 h and then transiently transfected with 30 μg of the histone H3.3 expression vector per dish. At 34 h after transfection, cells were induced using 20% FCS and collected for chromatin immunoprecipitation (ChIP) 8 h later. (B) NIH3T3 cells, treated as described in (A), were subjected to a ChIP assay using either the anti-haemagglutinin antibody (HA) or the anti-Flag antibody (Irr) as a control. The amounts of the dhfr (dihydrofolate reductase) promoter (DHFR-P), and P0 sequences were measured by quantitative PCR (Q-PCR). A representative experiment out of ten is shown. Ab, antibody. (C) Same as (B), except that NIH3T3 cells were transfected with either the H3.3–HA or the H3.1–HA expression vector. A representative experiment out of two is shown. (D) Same as (B), except that the amounts of the indicated sequences were measured by Q-PCR. A representative experiment out of three is shown.
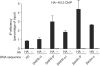
Figure 3
Analysis of histone H3.3 presence in stably expressing cells. NIH3T3 cells stably expressing tagged H3.3 were starved of serum for 48 h and then re-induced for 8 h with 20% FCS and subjected to a chromatin immunoprecipitation (ChIP) assay using the anti-haemagglutinin antibody (HA) or no antibody as a control. The amounts of the indicated sequences were measured by quantitative PCR. A representative experiment out of three is shown. Ab, antibody.
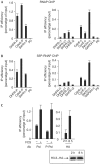
Figure 4
Deposition of H3.3 correlates with transcription. NIH3T3 cells were starved of serum for 48 h and then re-induced for 8 h with 20% FCS and subjected to a chromatin immunoprecipitation (ChIP) assay using an anti-pol II antibody (A) or an anti-S5-phosphorylated pol II antibody (B). The amounts of the indicated sequences were measured by quantitative PCR (Q-PCR). A representative experiment out of three is shown. (C) Left panel: NIH3T3 cells were starved of serum for 48 h and then re-induced or not, as indicated, with 20% FCS for 8 h. Right panel: NIH3T3 cells were transfected with the H3.3–haemagglutinin (HA) expression vector and re-induced for 2 or 8 h with 20% FCS, as indicated. Cells were then subjected to a ChIP assay using an anti-RNA polymerase II antibody (RNAP), anti-S5 phosphorylated RNAP antibody (S5P-RNAP; left panel) or an anti-HA antibody (right panel). The amount of dhfr (dihydrofolate reductase) promoter was measured by Q-PCR. A representative experiment out of three is shown. Also shown is a western blot monitoring H3.3–HA expression. Note that in the control we induced cells with serum for 2 h because exogenous histone expression is very low in uninduced cells (data not shown). Ab, antibody.
Similar articles
- Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication.
Jamai A, Imoberdorf RM, Strubin M. Jamai A, et al. Mol Cell. 2007 Feb 9;25(3):345-55. doi: 10.1016/j.molcel.2007.01.019. Mol Cell. 2007. PMID: 17289583 - Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter.
Nicolas E, Roumillac C, Trouche D. Nicolas E, et al. Mol Cell Biol. 2003 Mar;23(5):1614-22. doi: 10.1128/MCB.23.5.1614-1622.2003. Mol Cell Biol. 2003. PMID: 12588981 Free PMC article. - Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3.
Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Kraushaar DC, et al. Genome Biol. 2013;14(10):R121. doi: 10.1186/gb-2013-14-10-r121. Genome Biol. 2013. PMID: 24176123 Free PMC article. - Transcription-coupled H3.3 recycling: A link with chromatin states.
Delaney K, Almouzni G. Delaney K, et al. Semin Cell Dev Biol. 2023 Feb 15;135:13-23. doi: 10.1016/j.semcdb.2022.05.003. Epub 2022 May 18. Semin Cell Dev Biol. 2023. PMID: 35595602 Review. - H3.3 turnover: a mechanism to poise chromatin for transcription, or a response to open chromatin?
Huang C, Zhu B. Huang C, et al. Bioessays. 2014 Jun;36(6):579-84. doi: 10.1002/bies.201400005. Epub 2014 Apr 3. Bioessays. 2014. PMID: 24700556 Review.
Cited by
- The dark side of histones: genomic organization and role of oncohistones in cancer.
Amatori S, Tavolaro S, Gambardella S, Fanelli M. Amatori S, et al. Clin Epigenetics. 2021 Apr 7;13(1):71. doi: 10.1186/s13148-021-01057-x. Clin Epigenetics. 2021. PMID: 33827674 Free PMC article. Review. - Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome.
Wollmann H, Holec S, Alden K, Clarke ND, Jacques PÉ, Berger F. Wollmann H, et al. PLoS Genet. 2012;8(5):e1002658. doi: 10.1371/journal.pgen.1002658. Epub 2012 May 3. PLoS Genet. 2012. PMID: 22570629 Free PMC article. - H2A.B facilitates transcription elongation at methylated CpG loci.
Chen Y, Chen Q, McEachin RC, Cavalcoli JD, Yu X. Chen Y, et al. Genome Res. 2014 Apr;24(4):570-9. doi: 10.1101/gr.156877.113. Epub 2014 Jan 8. Genome Res. 2014. PMID: 24402521 Free PMC article. - Characterization of a Novel Chromatin Sorting Tool Reveals Importance of Histone Variant H3.3 in Contextual Fear Memory and Motor Learning.
McNally AG, Poplawski SG, Mayweather BA, White KM, Abel T. McNally AG, et al. Front Mol Neurosci. 2016 Feb 9;9:11. doi: 10.3389/fnmol.2016.00011. eCollection 2016. Front Mol Neurosci. 2016. PMID: 26903803 Free PMC article. - Histone exchange, chromatin structure and the regulation of transcription.
Venkatesh S, Workman JL. Venkatesh S, et al. Nat Rev Mol Cell Biol. 2015 Mar;16(3):178-89. doi: 10.1038/nrm3941. Epub 2015 Feb 4. Nat Rev Mol Cell Biol. 2015. PMID: 25650798 Review.
References
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 - PubMed
- Daury L, Trouche D (2003) Analysis of histone deposition on specific DNA elements in living mammalian cells. Biotechniques 35: 326–332 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases