Interleukin-21 receptor gene induction in human T cells is mediated by T-cell receptor-induced Sp1 activity - PubMed (original) (raw)
Interleukin-21 receptor gene induction in human T cells is mediated by T-cell receptor-induced Sp1 activity
Zheng Wu et al. Mol Cell Biol. 2005 Nov.
Abstract
Interleukin-21 (IL-21) plays important roles in regulating the immune response. IL-21 receptor (IL-21R) mRNA is expressed at a low level in human resting T cells but is rapidly induced by mitogenic stimulation. We now investigate the basis for IL21R gene regulation in T cells. We found that the -80 to -20 region critically regulates IL-21R promoter activity and corresponds to a major DNase I-hypersensitive site. Electrophoretic mobility shift assays, DNA affinity chromatography followed by mass spectrometry, and chromatin immunoprecipitation assays revealed that Sp1 binds to this region in vitro and in vivo. Moreover, mutation of the Sp1 motif markedly reduced IL-21R promoter activity, and Sp1 small interfering RNAs effectively diminished IL-21R expression in activated T cells. Interestingly, upon T-cell receptor (TCR) stimulation, T cells increased IL-21R expression and Sp1 protein levels while decreasing Sp1 phosphorylation. Moreover, phosphatase inhibitors that increased phosphorylation of Sp1 diminished IL-21R transcription. These data indicate that TCR-induced IL-21R expression is driven by TCR-mediated augmentation of Sp1 protein levels and may partly depend on the dephosphorylation of Sp1.
Figures
FIG. 1.
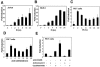
TCR-induced IL-21R expression in human T lymphocytes. Jurkat cells (A), Molt-3 cells (B), or purified human PB T cells (C) were cultured in the presence of 10 ng/ml PMA plus 1 μg/ml ionomycin for the indicated times. Human PB T cells were also cultured in the presence of 2 μg/ml anti-CD3 plus 1 μg/ml anti-CD28 antibody, as indicated (D). Human PB T cells were also incubated for 2 h with actinomycin D (10 μg/ml) or cycloheximide (10 μg/ml) with or without anti-CD3/anti-CD28 (E). Total RNA was isolated from cells, and first-strand cDNA was synthesized. IL-21R mRNA levels were quantitated by real-time PCR and normalized to the level of 18S rRNA. For each sample, the bar represents induction (_n_-fold) compared with no treatment. Values are means ± standard errors of the mean (SEM) of results from three experiments.
FIG. 2.
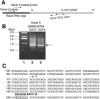
Mapping of a major human IL-21R transcription initiation site by 5′ RACE. (A) Locations of primers used in the 5′ RACE. IL21R GSPs 1, 2, and 3 are complementary to the regions from +805 to +828, +647 to +670, or +607 to +630, respectively (relative to the boundary of IL-21R 5′ cDNA) (35). RACER 5′ primer and RACER 5′ nested primer complementary to the RACER RNA oligonucleotide were provided by the GeneRACER kit. (B) Total RNA was isolated from human PB T cells treated with 2 μg/ml of anti-CD3 plus 1 μg/ml of anti-CD28 antibody for 2 h. The 5′ end of IL-21R was PCR amplified by RACER 5′ primer with GSP1 (product 1) or GSP2 (product 2). The nested PCR was performed by using nested primers and PCR product 1 (lane 2) and product 2 (lane 3). Lane 1 shows the molecular weight marker. Arrows indicate the bands cut from the gel for subcloning and sequencing. (C) Partial sequence of the 5′ regulatory region of the IL21R gene showing the major transcription initiation site (arrow) 243 nucleotides 5′ of exon 1a (open box).
FIG. 3.
Characterization of a DNase I-hypersensitive site in the human IL21R gene. (A) Schematic showing the location of a DNase I-hypersensitive site in the human IL21R gene. The thick bars represent the probes; the open box represents exon 1a. The major transcription initiation site mapped by 5′ RACE is indicated by the arrow. (B) Nuclei were isolated from Molt-3 cells cultured for 4 h with or without PI and then digested with DNase I as indicated. DNA was extracted, digested with either NdeI or BglI, and analyzed on a 0.6% agarose gel. The hypersensitive site was mapped by hybridizing NdeI-digested DNA with probe 1 (+6331 to +6754) and BglI-digested DNA with probe 2 (+1051 to +1352). (C) Nuclei isolated from human PB T cells cultured with or without anti-CD3 and anti-CD28 were digested with DNase I. The hypersensitive site was mapped by hybridizing BglI-digested DNA with probe 2. (D) Acetylation levels of the IL21R gene on chromosome 16 in resting primary human T cells (upper panel) or anti-CD3 and anti-CD28 antibody-activated (24 h) primary T cells (lower panel) were analyzed by GMAT. The genome-wide data are posted at the website
http://dir.nhlbi.nih.gov/labs/lmi/zhao/epigenome/G&D2005.htm
. The retrieved region includes the promoter and the first intron of the IL21R gene, as indicated by the scheme at the bottom. (E) Nuclei isolated from human PB T cells treated with anti-CD3 and anti-CD28 for 4 h or not treated were briefly digested with BstEII. The purified genomic DNA was digested to completion with StuI. The cleavage sites were detected by linker ligation-mediated PCR with the antisense IL-21R GSP. The data were quantified by PhosphorImager. The intensity of the BstEII band was normalized for the intensity of the StuI band. The bars represent induction of BstEII accessibility (_n_-fold) compared with no treatment. Values are means ± SEM of results from three experiments. Locations of restriction enzymes and primers for PCR are shown at the top.
FIG. 4.
The −80 to −20 region is important for IL-21R promoter activity in response to TCR activation and forms complexes with nuclear factors. (A) Luciferase (Luc) reporter construct schematics are on the left. These were transfected into Molt-3 cells that were not stimulated or stimulated with PI. Luciferase activity was normalized with renilla luciferase activity (relative light units, RLU). Shown are means ± SEM of results from three experiments. (B) Nuclear extracts from PI-treated (4 h) or untreated human PB T cells were incubated with the indicated IL-21R promoter probes and subjected to EMSA. Two DNA-binding protein complexes, C1 and C2, are indicated. (C) EMSA with the −61 to −32 probe and nuclear extract from untreated or PI-treated human PB T cells. In lane 3, a 100-fold molar excess of unlabeled −61 to −32 oligonucleotide was preincubated with nuclear extract before adding the probe.
FIG. 5.
Characterization of the TGGGCG motif in the 5′ regulatory region of IL-21R responsible for TCR-induced activity. (A) Sequence of the wild-type (WT) and mutant (Mut) −61 to −32 IL-21R probes used in EMSAs. (B) EMSAs with WT or Mut probes and nuclear extract from human PB T cells treated with PI for 4 h or not treated. (C) Transient transfection of IL-21R promoter reporter constructs in Molt-3 cells and mouse splenic T cells. Luciferase (Luc) reporter construct schematics are on the left. Mut5 and Mut6 are shown in panel A. The luciferase constructs were transfected into Molt-3 cells followed by no stimulation or stimulation with PI or transfected into preactivated mouse splenic T cells that were then not treated or stimulated with anti-CD3/CD28. Luciferase activity was measured and normalized with Renilla luciferase activity (relative light units, RLU). Values are means ± SEM of results from three experiments.
FIG. 6.
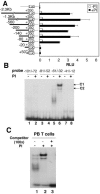
Sp1 and Sp3 bind to the GC motif in the IL-21R promoter in vitro. (A) Proteins purified by DNA affinity chromatography. Nuclear extracts from PI-stimulated (4 h) human PB T cells were subjected to DNA affinity chromatography. Eluates from WT or mutant (Mut5, as in Fig. 5A) DNA-conjugated beads were resolved by SDS-PAGE and silver stained. LC-MS and MALDI-TOF mass spectrometry were conducted on the 95-, 80-, and 40-kDa bands (arrows). The 95-kDa band is Sp1. (B) Sequence comparison of human and mouse IL-21R promoter-proximal region showed a conserved GGGCGGGGC motif. (C) EMSA using WT probe and recombinant Sp1, Sp3, or nuclear extracts (NE) from human PB T cells treated with PI or not treated. Antibodies to Sp1, Sp2, Sp3, Sp4, or unlabeled oligonucleotides containing the Sp1 consensus sequence at 25-, 50-, or 100-fold molar excess were preincubated with nuclear extract before adding WT probe, as indicated. (D) Nuclear extracts (15 μg) from anti-CD3-plus-anti-CD28-activated human PB T cells or mouse splenic T cells were blotted with anti-Sp1 or γ-tubulin (control). (E) Nuclear extracts (15 μg) were blotted with anti-Sp3 or γ-tubulin.
FIG. 7.
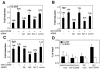
Binding of Sp1 to the IL-21R promoter is essential for TCR-induced IL-21R expression. Suppression of IL-21R expression in human primary T cells by siRNA targeting of Sp1 is also shown. Human PB T cells were transfected with 100 nM siRNAs along with pEYFP-N1 and were cultured for 36 h. YFP+ cells were sorted and stimulated with anti-CD3 and anti-CD28 for 2 h. Total RNA was extracted and analyzed for IL-21R (A), Sp1 (B), and Sp3 (C) expression by quantitative real-time PCR. IL-21R, Sp1, and Sp3 expressions were normalized to the level of expression of 18S rRNA. For each sample, the bar represents the induction (_n_-fold) compared to no TCR activation. Values are means ± SEM of results from three experiments. (D) In vivo binding of Sp1 to the IL-21R promoter in human primary T cells by chromatin immunoprecipitation. Human PB T cells were cultured with or without anti-CD3 and anti-CD28 for 2 h. Formaldehyde-cross-linked chromatin was immunoprecipitated with preimmune rabbit serum or with anti-Sp1. Real-time PCR was performed to quantitate the DNA fragment containing the Sp1 binding site in the IL-21R promoter. β-Actin was a control. Values are means ± SEM of results from three experiments.
FIG. 8.
Dephosphorylation of Sp1 is essential for TCR activation-induced IL-21R expression. Human PB T cells were not stimulated or stimulated with anti-CD3 and anti-CD28 for 2 h. Half of the cells were coincubated with 10 nM calyculin A or 0.8 μM okadaic acid. Total RNA and nuclear extract were isolated from these cells. (A) Suppression of TCR-induced IL-21R mRNA expression by calyculin A or okadaic acid treatment. Total RNA was transcribed to first-strand cDNA, and IL-21R mRNA levels were quantitated by real-time PCR and normalized to the level of 18S rRNA. The bar represents the induction (_n_-fold) compared with no treatment. Values are means ± SEM of results from three experiments. (B) Calyculin A or okadaic acid treatment diminished the Sp1 DNA-binding activity. Nuclear extracts were incubated with the −61 to −32 probe of IL-21R promoter and subjected to EMSA. Different donors were used by the calyculin A and okadaic acid experiments, presumably explaining the difference in the levels of Sp1. (C) Calyculin A or okadaic acid treatment inhibited Sp1 dephosphorylation. Fifteen micrograms of the nuclear extract was subjected to SDS-PAGE on 8% Novex Tris-glycine gel or 8% NuPAGE Tris-acetate gel, transferred to an Immobilon-P membrane, and blotted with anti-Sp1 or γ-tubulin.
Similar articles
- The gene for interleukin-21 receptor is the partner of BCL6 in t(3;16)(q27;p11), which is recurrently observed in diffuse large B-cell lymphoma.
Ueda C, Akasaka T, Kurata M, Maesako Y, Nishikori M, Ichinohasama R, Imada K, Uchiyama T, Ohno H. Ueda C, et al. Oncogene. 2002 Jan 17;21(3):368-76. doi: 10.1038/sj.onc.1205099. Oncogene. 2002. PMID: 11821949 - Laminin reduces expression of the human alpha6 integrin subunit gene by altering the level of the transcription factors Sp1 and Sp3.
Gaudreault M, Vigneault F, Leclerc S, Guérin SL. Gaudreault M, et al. Invest Ophthalmol Vis Sci. 2007 Aug;48(8):3490-505. doi: 10.1167/iovs.07-0016. Invest Ophthalmol Vis Sci. 2007. PMID: 17652716 - IFN-alpha regulates IL-21 and IL-21R expression in human NK and T cells.
Strengell M, Julkunen I, Matikainen S. Strengell M, et al. J Leukoc Biol. 2004 Aug;76(2):416-22. doi: 10.1189/jlb.1003488. Epub 2004 Jun 3. J Leukoc Biol. 2004. PMID: 15178704 - Interleukin 21: a key player in lymphocyte maturation.
Nutt SL, Brady J, Hayakawa Y, Smyth MJ. Nutt SL, et al. Crit Rev Immunol. 2004;24(4):239-50. doi: 10.1615/critrevimmunol.v24.i4.20. Crit Rev Immunol. 2004. PMID: 15588224 Review. - Transcriptional regulation by post-transcriptional modification--role of phosphorylation in Sp1 transcriptional activity.
Chu S. Chu S. Gene. 2012 Oct 15;508(1):1-8. doi: 10.1016/j.gene.2012.07.022. Epub 2012 Jul 24. Gene. 2012. PMID: 22835698 Review.
Cited by
- Interaction of papillomavirus E2 protein with the Brm chromatin remodeling complex leads to enhanced transcriptional activation.
Kumar RA, Naidu SR, Wang X, Imbalzano AN, Androphy EJ. Kumar RA, et al. J Virol. 2007 Mar;81(5):2213-20. doi: 10.1128/JVI.01746-06. Epub 2006 Dec 6. J Virol. 2007. PMID: 17151122 Free PMC article. - Differential effects of IL-21 and IL-15 on perforin expression, lysosomal degranulation, and proliferation in CD8 T cells of patients with human immunodeficiency virus-1 (HIV).
White L, Krishnan S, Strbo N, Liu H, Kolber MA, Lichtenheld MG, Pahwa RN, Pahwa S. White L, et al. Blood. 2007 May 1;109(9):3873-80. doi: 10.1182/blood-2006-09-045278. Epub 2006 Dec 27. Blood. 2007. PMID: 17192392 Free PMC article. Clinical Trial. - Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus.
McGuire HM, Vogelzang A, Hill N, Flodström-Tullberg M, Sprent J, King C. McGuire HM, et al. Proc Natl Acad Sci U S A. 2009 Nov 17;106(46):19438-43. doi: 10.1073/pnas.0903561106. Epub 2009 Oct 30. Proc Natl Acad Sci U S A. 2009. PMID: 19880748 Free PMC article. - Interleukin-21 and cellular activation concurrently induce potent cytotoxic function and promote antiviral activity in human CD8 T cells.
Parmigiani A, Pallin MF, Schmidtmayerova H, Lichtenheld MG, Pahwa S. Parmigiani A, et al. Hum Immunol. 2011 Feb;72(2):115-23. doi: 10.1016/j.humimm.2010.10.015. Epub 2010 Oct 25. Hum Immunol. 2011. PMID: 20977918 Free PMC article. - The Yin and Yang of interleukin-21 in allergy, autoimmunity and cancer.
Spolski R, Leonard WJ. Spolski R, et al. Curr Opin Immunol. 2008 Jun;20(3):295-301. doi: 10.1016/j.coi.2008.02.004. Epub 2008 Jun 12. Curr Opin Immunol. 2008. PMID: 18554883 Free PMC article. Review.
References
- Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. - PubMed
- Asao, H., C. Okuyama, S. Kumaki, N. Ishii, S. Tsuchiya, D. Foster, and K. Sugamura. 2001. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J. Immunol. 167:1-5. - PubMed
- Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188:143-160. - PubMed
- Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195:27-38. - PubMed
- Briggs, M. R., J. T. Kadonaga, S. P. Bell, and R. Tjian. 1986. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234:47-52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources