The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions - PubMed (original) (raw)
The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions
Klaus F Hartlepp et al. Mol Cell Biol. 2005 Nov.
Abstract
The chromatin accessibility complex (CHRAC) is an abundant, evolutionarily conserved nucleosome remodeling machinery able to catalyze histone octamer sliding on DNA. CHRAC differs from the related ACF complex by the presence of two subunits with molecular masses of 14 and 16 kDa, whose structure and function were not known. We determined the structure of Drosophila melanogaster CHRAC14-CHRAC16 by X-ray crystallography at 2.4-angstroms resolution and found that they dimerize via a variant histone fold in a typical handshake structure. In further analogy to histones, CHRAC14-16 contain unstructured N- and C-terminal tail domains that protrude from the handshake structure. A dimer of CHRAC14-16 can associate with the N terminus of ACF1, thereby completing CHRAC. Low-affinity interactions of CHRAC14-16 with DNA significantly improve the efficiency of nucleosome mobilization by limiting amounts of ACF. Deletion of the negatively charged C terminus of CHRAC16 enhances DNA binding 25-fold but leads to inhibition of nucleosome sliding, in striking analogy to the effect of the DNA chaperone HMGB1 on nucleosome sliding. The presence of a surface compatible with DNA interaction and the geometry of an H2A-H2B heterodimer may provide a transient acceptor site for DNA dislocated from the histone surface and therefore facilitate the nucleosome remodeling process.
Figures
FIG. 1.
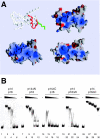
Structure of the CHRAC p14-p16 heterodimer. (A) Alignment of Drosophila CHRAC14 and CHRAC16 with human and mouse homologues, NFYB and NFYC, subunits of the human transcription factor NFY, and Xenopus histones H2A and H2B. Conserved or conservatively substituted residues within the p14-p16 families and compared to NFYB-NFYC are depicted with a yellow background. In addition, residues conserved or conservatively substituted in H2A-H2B compared to p14-p16 and NFYB-NFYC are also shown with a yellow background. Secondary structure elements in the p14 and p16 subunits were determined by manual inspection. The noncanonical p16 helix αC is shown in light blue. Secondary structure elements of NFYB-NFYC and H2A-H2B are depicted as published previously (33, 41). Disordered regions present in the initial constructs are represented by broken lines and are not included in the final model. H2A-H2B residues forming hydrogen bonds with nucleosomal DNA are shown in boxes. Intrachain salt bridges conserved between p14 and NFYB (red), but not between p16 and NFYC (blue), are indicated. (B) Ribbon representation of the p14-p16 heterodimer. CHRAC14 and CHRAC16 are depicted in red and blue, respectively. The color code for the two CHRAC subunits is kept throughout the figures. Figure 1B (see also Fig. S1 and S3 in the supplemental material) was produced with program Ribbons (9).
FIG. 2.
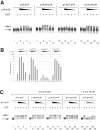
p14-p16 interacts with the N terminus of ACF1. (A) ACF1 derivatives used in this study. Top panel: baculovirus-encoded, _myc_-tagged ACF1 derivatives; bottom panel: in vitro-translated ACF1 derivatives. Interaction with p14-p16, as determined by the results shown in panels B and D, is indicated to the right: +, interaction; −, no interaction. (B) FLAG affinity purification of complexes from whole-cell extracts of SF9 cells coinfected with p14FLAG-HISp16 and _myc_-tagged ACF1 variants (shown in panel A). The Western blot was probed with anti-myc antibody. In the mock infection (lane 1), the cells were transfected with p14FLAG-HISp16 alone. Bands corresponding to the ACF1 constructs in the input are marked with asterisks. (C) Coomassie-stained 15% polyacrylamide gel of glutathione-Sepharose beads loaded with purified GST, GSTp14, and GSTp14-HISp16 (3.75 μg/lane). (D) GST pull down of in vitro-translated ACF1 constructs (shown in panel A). Top panel: 5% of input; bottom panel: pull down with recombinant GSTp14-HISp16 heterodimer.
FIG. 3.
ACF-catalyzed nucleosome sliding is enhanced by p14-p16. Six-nanomolar radiolabeled end-positioned nucleosomes were incubated with approximately 300 pM, 90 pM, and 30 pM ACF, and decreasing amounts of p14-p16 (8, 4, 2, and 1 μM) were added to the reaction mixtures. With 300 pM ACF, sliding is maximal and no further enhancement due to p14-p16 is seen (lanes 2 to 6). Nucleosomes do not slide in the absence of ATP (lanes 9 to 13). Migration of the nucleosomes is not affected by p14-p16 alone (lane 7 and 14). +, present; −, absent. The bands correspond to (from the top) centrally positioned nucleosome, end-positioned nucleosome, and free DNA fragment, as schematically indicated to the left.
FIG. 4.
The p14-p16 sliding enhancement is mediated by the ACF1 WAC domain. (A) Protein titrations. Nucleosome sliding is catalyzed by 300, 75, and 37.5 pM concentrations of either ACF or ACFΔWAC (decreasing shading of bar) in the presence of decreasing amounts of p14-p16 (8, 4, 2, and 1 μM) as indicated. (B) Time course. Reactions were performed with concentrations of ACF and ACFΔWAC complex (approximately 180 pM) which do not suffice to slide nucleosomes in the absence or presence of p14-p16 (approximately 8 μM). Time points are taken after 0, 2, 5, 10, 20, and 45 min. −, absent. Labeling is as described for Fig. 3.
FIG. 5.
Role of N- and C-terminal tails of p14-p16 for ACF1 binding. (A) Summary of p14-p16 derivatives. (B) Coomassie-stained 15% polyacrylamide gel of glutathione-Sepharose beads loaded with 3.75 μg of purified recombinant GSTp14-p16 derivatives as indicated. (C) GST pull down of in vitro-translated ACF1 constructs (shown in Fig. 1C). Lane 1, 5% of input. The p14-p16 derivatives used for the pull down are indicated above the lanes. The in vitro-translated ACF1 derivatives assayed for interaction are indicated to the right.
FIG. 6.
DNA binding by CHRAC p14-p16. (A) Surface charge distribution of p14-p16 compared to that of NFYB-NFYC and H2A-H2B heterodimers calculated with GRASP (38). Negative and positive potentials (±15 kB T [kB, Boltzmann constant; T, temperature]) are depicted in red and blue, respectively. The orientation of the three protein pairs is identical and corresponds to that shown for the p14-p16 worm model. Helix α1 of CHRAC16 was modeled onto NFYC and is depicted in red (compare also to Fig. S1 in the supplemental material). (B) EMSA with wt p14-p16 and p14-p16 deletion mutants. Six-nanomolar concentrations of radiolabeled 248-bp DNA were incubated with 20, 5, 2, 0.5, and 0.2 μM of the respective p14-p16 derivatives before complexes were resolved on a native polyacrylamide gel. An autoradiography of the dried gel is shown. −, absent.
FIG. 7.
Effect of p14-p16 tails on nucleosome sliding. (A) Deletion of the p14 C terminus leads to loss of sliding enhancement. Sliding reaction mixtures contained 6 nM concentrations of end-positioned nucleosome, 37.5 pM ACF, and 2, 1, 0.5, and 0.25 μM concentrations of intact p14-p16 (lanes 3 to 6), p14ΔN-p16 (lanes 9 to 12), p14ΔC-p16 (lanes 15 to 18), or p14-p16ΔN (lanes 21 to 24). (B) Quantification of nucleosome sliding. The percentage of nucleosomes that had been moved from the end to the center position was determined using a phosphorimager (Fuji). The graph shows the average sliding results from two independent experiments. (C) Deletion of the p16 C terminus inhibits ACF-catalyzed nucleosome sliding. Sliding reactions contained 6 nM concentrations of end-positioned nucleosome, 0.3 nM ACF, and different p14-p16 derivatives (6, 3, 1.5, and 0.75 μM), with the exception of p14-p16ΔC, for which concentrations of 1.5, 0.75, 0.38, and 0.19 μM were used (lanes 26 to 30). +, present; −, absent. Labeling is as described for Fig. 3.
Similar articles
- The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF.
Kukimoto I, Elderkin S, Grimaldi M, Oelgeschläger T, Varga-Weisz PD. Kukimoto I, et al. Mol Cell. 2004 Jan 30;13(2):265-77. doi: 10.1016/s1097-2765(03)00523-9. Mol Cell. 2004. PMID: 14759371 - Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC).
Corona DF, Eberharter A, Budde A, Deuring R, Ferrari S, Varga-Weisz P, Wilm M, Tamkun J, Becker PB. Corona DF, et al. EMBO J. 2000 Jun 15;19(12):3049-59. doi: 10.1093/emboj/19.12.3049. EMBO J. 2000. PMID: 10856248 Free PMC article. - HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins.
Poot RA, Dellaire G, Hülsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. Poot RA, et al. EMBO J. 2000 Jul 3;19(13):3377-87. doi: 10.1093/emboj/19.13.3377. EMBO J. 2000. PMID: 10880450 Free PMC article. - DNA binding within the nucleosome core.
Luger K, Richmond TJ. Luger K, et al. Curr Opin Struct Biol. 1998 Feb;8(1):33-40. doi: 10.1016/s0959-440x(98)80007-9. Curr Opin Struct Biol. 1998. PMID: 9519294 Review. - Histone chaperones and nucleosome assembly.
Akey CW, Luger K. Akey CW, et al. Curr Opin Struct Biol. 2003 Feb;13(1):6-14. doi: 10.1016/s0959-440x(03)00002-2. Curr Opin Struct Biol. 2003. PMID: 12581654 Review.
Cited by
- Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits.
Calvenzani V, Testoni B, Gusmaroli G, Lorenzo M, Gnesutta N, Petroni K, Mantovani R, Tonelli C. Calvenzani V, et al. PLoS One. 2012;7(8):e42902. doi: 10.1371/journal.pone.0042902. Epub 2012 Aug 17. PLoS One. 2012. PMID: 22912760 Free PMC article. - CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold.
Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. Nishino T, et al. Cell. 2012 Feb 3;148(3):487-501. doi: 10.1016/j.cell.2011.11.061. Cell. 2012. PMID: 22304917 Free PMC article. - Importin 13 mediates nuclear import of histone fold-containing chromatin accessibility complex heterodimers.
Walker P, Doenecke D, Kahle J. Walker P, et al. J Biol Chem. 2009 Apr 24;284(17):11652-62. doi: 10.1074/jbc.M806820200. Epub 2009 Feb 14. J Biol Chem. 2009. PMID: 19218565 Free PMC article. - NF-Y substitutes H2A-H2B on active cell-cycle promoters: recruitment of CoREST-KDM1 and fine-tuning of H3 methylations.
Gatta R, Mantovani R. Gatta R, et al. Nucleic Acids Res. 2008 Nov;36(20):6592-607. doi: 10.1093/nar/gkn699. Epub 2008 Oct 21. Nucleic Acids Res. 2008. PMID: 18940868 Free PMC article. Retracted. - Quantitative determination of binding of ISWI to nucleosomes and DNA shows allosteric regulation of DNA binding by nucleotides.
Al-Ani G, Briggs K, Malik SS, Conner M, Azuma Y, Fischer CJ. Al-Ani G, et al. Biochemistry. 2014 Jul 15;53(27):4334-45. doi: 10.1021/bi500224t. Epub 2014 Jun 30. Biochemistry. 2014. PMID: 24898734 Free PMC article.
References
- Becker, P. B., and W. Hörz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. - PubMed
- Birck, C., O. Poch, C. Romier, M. Ruff, G. Mengus, A. C. Lavigne, I. Davidson, and D. Moras. 1998. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94:239-249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases