Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection - PubMed (original) (raw)
Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection
John D F Hale et al. Appl Environ Microbiol. 2005 Nov.
Abstract
Streptococcus mutans UA159, the genome sequence reference strain, exhibits nonlantibiotic mutacin activity. In this study, bioinformatic and mutational analyses were employed to demonstrate that the antimicrobial repertoire of strain UA159 includes mutacin IV (specified by the nlm locus) and a newly identified bacteriocin, mutacin V (encoded by SMU.1914c).
Figures
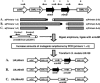
FIG. 1.
Allelic replacement, by PCR ligation mutagenesis (13), of genes (nlmAB) encoding the peptides comprising mutacin IV. Other putative bacteriocin-encoding genes investigated in this study (see Fig. 2) were also inactivated using this strategy. For clarity, the loci are not drawn to scale. The PCR primers shown are numbered as follows: 1, NlmABUpF; 2, NlmAUpR; 3, NlmADwF; 4, NlmBUpR; 5, NlmBDwF; 6, NlmABDwR. The filled and unfilled circles refer to EcoRI and PstI restriction sites, respectively. The source of ermAM was pSLER1 (pSL1190 [4] containing ermAM cloned into the NdeI site; laboratory collection) and ermAM was always inserted in the same transcriptional orientation as the gene of interest. The locations of the ErmInv PCR primers used to confirm each mutation are also shown. adhE, putative alcohol-acetaldehyde dehydrogenase; nlmA and nlmB, prepeptides of NlmA* and NlmB*, respectively; SMU.149, putative transposase; SMU.152, hypothetical protein.
FIG. 2.
Genomic organization of the open reading frames (ORFs; filled pentagons) encoding potential nonlantibiotic mutacins investigated in this study. For simplicity, the loci are not drawn to scale. ORFs designated by their GenBank locus tags (e.g., SMU.283) encode hypothetical proteins. Note that SMU.282 does not exist. The translational orientation of each ORF is also indicated. nlmTE, ABC transporter required for export of nonlantibiotic mutacins (10); tif2, translation initiation factor 2; rbfA, ribosome binding factor A; copY, putative transcriptional regulator; copAZ, components of a copper transport system; tnp, putative transposase of IS_Smu1_ (1); cslAB, ABC transport system required for natural transformation (10, 18); comCDE, signal transduction system essential for development of natural competence (8); dedA, putative membrane-associated protein.
Similar articles
- Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans.
Hale JD, Heng NC, Jack RW, Tagg JR. Hale JD, et al. J Bacteriol. 2005 Jul;187(14):5036-9. doi: 10.1128/JB.187.14.5036-5039.2005. J Bacteriol. 2005. PMID: 15995224 Free PMC article. - Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8.
Robson CL, Wescombe PA, Klesse NA, Tagg JR. Robson CL, et al. Microbiology (Reading). 2007 May;153(Pt 5):1631-1641. doi: 10.1099/mic.0.2006/003756-0. Microbiology (Reading). 2007. PMID: 17464078 - The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV.
Qi F, Chen P, Caufield PW. Qi F, et al. Appl Environ Microbiol. 2001 Jan;67(1):15-21. doi: 10.1128/AEM.67.1.15-21.2001. Appl Environ Microbiol. 2001. PMID: 11133423 Free PMC article. - LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component.
Merritt J, Kreth J, Shi W, Qi F. Merritt J, et al. Mol Microbiol. 2005 Aug;57(4):960-9. doi: 10.1111/j.1365-2958.2005.04733.x. Mol Microbiol. 2005. PMID: 16091037 - The mutacins of Streptococcus mutans: regulation and ecology.
Merritt J, Qi F. Merritt J, et al. Mol Oral Microbiol. 2012 Apr;27(2):57-69. doi: 10.1111/j.2041-1014.2011.00634.x. Epub 2011 Dec 23. Mol Oral Microbiol. 2012. PMID: 22394465 Free PMC article. Review.
Cited by
- Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation.
Sztajer H, Lemme A, Vilchez R, Schulz S, Geffers R, Yip CY, Levesque CM, Cvitkovitch DG, Wagner-Döbler I. Sztajer H, et al. J Bacteriol. 2008 Jan;190(1):401-15. doi: 10.1128/JB.01086-07. Epub 2007 Nov 2. J Bacteriol. 2008. PMID: 17981981 Free PMC article. - Competence-dependent endogenous DNA rearrangement and uptake of extracellular DNA give a natural variant of Streptococcus mutans without biofilm formation.
Narisawa N, Kawarai T, Suzuki N, Sato Y, Ochiai K, Ohnishi M, Watanabe H, Senpuku H. Narisawa N, et al. J Bacteriol. 2011 Oct;193(19):5147-54. doi: 10.1128/JB.05240-11. Epub 2011 Jul 29. J Bacteriol. 2011. PMID: 21804005 Free PMC article. - Fighting biofilms with lantibiotics and other groups of bacteriocins.
Mathur H, Field D, Rea MC, Cotter PD, Hill C, Ross RP. Mathur H, et al. NPJ Biofilms Microbiomes. 2018 Apr 19;4:9. doi: 10.1038/s41522-018-0053-6. eCollection 2018. NPJ Biofilms Microbiomes. 2018. PMID: 29707229 Free PMC article. Review. - Phenotypic characterization of the foldase homologue PrsA in Streptococcus mutans.
Guo L, Wu T, Hu W, He X, Sharma S, Webster P, Gimzewski JK, Zhou X, Lux R, Shi W. Guo L, et al. Mol Oral Microbiol. 2013 Apr;28(2):154-65. doi: 10.1111/omi.12014. Epub 2012 Dec 13. Mol Oral Microbiol. 2013. PMID: 23241367 Free PMC article. - Quantitative Proteomics Uncovers the Interaction between a Virulence Factor and Mutanobactin Synthetases in Streptococcus mutans.
Rainey K, Wilson L, Barnes S, Wu H. Rainey K, et al. mSphere. 2019 Sep 25;4(5):e00429-19. doi: 10.1128/mSphere.00429-19. mSphere. 2019. PMID: 31554721 Free PMC article.
References
- Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. K. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. - PMC - PubMed
- Balakrishnan, M., R. S. Simmonds, A. Carne, and J. R. Tagg. 2000. Streptococcus mutans strain N produces a novel low molecular mass non-lantibiotic bacteriocin. FEMS Microbiol. Lett. 183:165-169. - PubMed
- Brosius, J. 1989. Superpolylinkers in cloning and expression vectors. DNA 8:759-777. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources