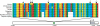Ancient and recent positive selection transformed opioid cis-regulation in humans - PubMed (original) (raw)
Ancient and recent positive selection transformed opioid cis-regulation in humans
Matthew V Rockman et al. PLoS Biol. 2005 Dec.
Abstract
Changes in the cis-regulation of neural genes likely contributed to the evolution of our species' unique attributes, but evidence of a role for natural selection has been lacking. We found that positive natural selection altered the cis-regulation of human prodynorphin, the precursor molecule for a suite of endogenous opioids and neuropeptides with critical roles in regulating perception, behavior, and memory. Independent lines of phylogenetic and population genetic evidence support a history of selective sweeps driving the evolution of the human prodynorphin promoter. In experimental assays of chimpanzee-human hybrid promoters, the selected sequence increases transcriptional inducibility. The evidence for a change in the response of the brain's natural opioids to inductive stimuli points to potential human-specific characteristics favored during evolution. In addition, the pattern of linked nucleotide and microsatellite variation among and within modern human populations suggests that recent selection, subsequent to the fixation of the human-specific mutations and the peopling of the globe, has favored different prodynorphin cis-regulatory alleles in different parts of the world.
Figures
Figure 1. Divergence of the 68-bp Element in Humans
Arrows indicate five differences fixed on the human lineage. The asterisk indicates a site that varies among human repeats. In the sample of 74 human haplotypes, all one-repeat and most two-repeat alleles bear G at this site. Complete haplotype data are given in Table S1. Below, schematic of the study region showing the position of the element and the non-coding first exon with respect to the start of transcription.
Figure 2. The Human 68-bp Element Increases Induced PDYN Expression
We tested four 3-kb constructs, encompassing the region shown in Figure 1. (A) The human and chimpanzee constructs differ at the sites indicated by vertical bars. The two chimeric constructs incorporated the human 68-bp element or the human DRE (DREAM binding site) into the chimpanzee construct. (B and C) Panels show luciferase activity for each construct (± SEM, five to seven transfections), standardized to that observed for promoterless luciferase vectors (white bars), in SH-SY5Y and JAR cells, with and without added caffeine, which causes the release of intracellular Ca2+ and the release of DREAM from the DRE.
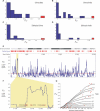
Figure 3. Elevated Differentiation at PDYN
(A–D) In four pairwise comparisons, FST at the PDYN 68-bp element (red) is markedly elevated above the FST estimated from 18 candidate neutral markers (blue) typed in the same individuals. (E) Genetic differentiation between European- and Chinese-Americans, measured as a 15-SNP running FST average, for the entire p-arm of Chromosome 20. PDYN falls under a large FST peak (shaded), high above the arm average (red line). The RefSeq and chromosome band annotation is from the University of California, Santa Cruz Human Genome Browser (hg17),
[79]. Perlegen SNP positions were matched to the hg17 assembly by the UCSC LiftOver utility. (F) A finer-scale sliding window analysis shows that the region of elevated FST includes only two genes, PDYN and STK35, shown according to their RefSeq annotations. (G) FST as a function of expected global heterozygosity. Red triangles represent the 52 SNPs in the Perlegen dataset in the 170-kb interval bounded by the 3′ ends of PDYN and STK35. The contours define the genome-wide density of FST conditioned on heterozygosity; for each heterozygosity, the lines represent the FST of SNPs in the specified FST percentile.
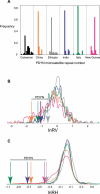
Figure 4. Altered Variation at the PDYN Microsatellite
(A) The allele frequency distribution of the PDYN microsatellite for six populations. The most common allele has 18 CA repeats in each population except Papua New Guinea, where the 22-repeat allele is most common; the overall range is 13 to 27 repeats. The distributions show a reduction in allelic variation outside of the Cameroon population. (B) The empirical probability density of lnRV for a panel of genomically distributed microsatellites is plotted for each population, using panel A as the color key. The distributions are based on 193 microsatellite loci for Ethiopia and 377 loci for the other populations. For clarity, a single negative outlier from the New Guinea population has been omitted from the figure. The arrows indicate lnRV of the PDYN microsatellite for each population, in the left tails of the distributions, indicating a locus-specific reduction in repeat-number variance. (C) The empirical probability density for lnRH. Again, the PDYN microsatellite exhibits significantly negative lnRH values, indicating a locus-specific reduction in heterozygosity at PDYN in the non-West African populations.
Similar articles
- Identification of multiple DNA elements regulating basal and protein kinase A-induced transcriptional expression of the rat prodynorphin gene.
Douglass J, McKinzie AA, Pollock KM. Douglass J, et al. Mol Endocrinol. 1994 Mar;8(3):333-44. doi: 10.1210/mend.8.3.8015551. Mol Endocrinol. 1994. PMID: 8015551 - Genetics. Expression of endorphin gene favored in human evolution.
Balter M. Balter M. Science. 2005 Nov 25;310(5752):1257. doi: 10.1126/science.310.5752.1257. Science. 2005. PMID: 16311304 No abstract available. - Multiple Functional Variants in cis Modulate PDYN Expression.
Babbitt CC, Silverman JS, Haygood R, Reininga JM, Rockman MV, Wray GA. Babbitt CC, et al. Mol Biol Evol. 2010 Feb;27(2):465-79. doi: 10.1093/molbev/msp276. Epub 2009 Nov 12. Mol Biol Evol. 2010. PMID: 19910384 - Trends in the evolution of the proenkephalin and prodynorphin genes in gnathostomes.
Khalap A, Bagrosky B, Lecaude S, Youson J, Danielson P, Dores RM. Khalap A, et al. Ann N Y Acad Sci. 2005 Apr;1040:22-37. doi: 10.1196/annals.1327.003. Ann N Y Acad Sci. 2005. PMID: 15891003 Review. - Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain.
Nosova O, Bazov I, Karpyak V, Hallberg M, Bakalkin G. Nosova O, et al. Molecules. 2021 Jun 7;26(11):3458. doi: 10.3390/molecules26113458. Molecules. 2021. PMID: 34200173 Free PMC article. Review.
Cited by
- The evolution of mammalian gene families.
Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. Demuth JP, et al. PLoS One. 2006 Dec 20;1(1):e85. doi: 10.1371/journal.pone.0000085. PLoS One. 2006. PMID: 17183716 Free PMC article. - Accelerated evolution at chaperone promoters among Antarctic notothenioid fishes.
Bogan SN, Place SP. Bogan SN, et al. BMC Evol Biol. 2019 Nov 6;19(1):205. doi: 10.1186/s12862-019-1524-y. BMC Evol Biol. 2019. PMID: 31694524 Free PMC article. - Population structure in a comprehensive genomic data set on human microsatellite variation.
Pemberton TJ, DeGiorgio M, Rosenberg NA. Pemberton TJ, et al. G3 (Bethesda). 2013 May 20;3(5):891-907. doi: 10.1534/g3.113.005728. G3 (Bethesda). 2013. PMID: 23550135 Free PMC article. - Coevolution within and between regulatory loci can preserve promoter function despite evolutionary rate acceleration.
Barrière A, Gordon KL, Ruvinsky I. Barrière A, et al. PLoS Genet. 2012 Sep;8(9):e1002961. doi: 10.1371/journal.pgen.1002961. Epub 2012 Sep 20. PLoS Genet. 2012. PMID: 23028368 Free PMC article. - Tempo and mode in the endocannaboinoid system.
McPartland JM, Norris RW, Kilpatrick CW. McPartland JM, et al. J Mol Evol. 2007 Sep;65(3):267-76. doi: 10.1007/s00239-007-9004-1. Epub 2007 Aug 4. J Mol Evol. 2007. PMID: 17676365
References
- Stedman HH, Kozyak BW, Nelson A, Thesier DM, Su LT, et al. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature. 2004;428:415–418. - PubMed
- Winter H, Langbein L, Krawczak M, Cooper DN, Jave-Suarez LF, et al. Human type I hair keratin pseudogene phihHaA has functional orthologs in the chimpanzee and gorilla: Evidence for recent inactivation of the human gene after the Pan-Homo divergence. Hum Genet. 2001;108:37–42. - PubMed
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources